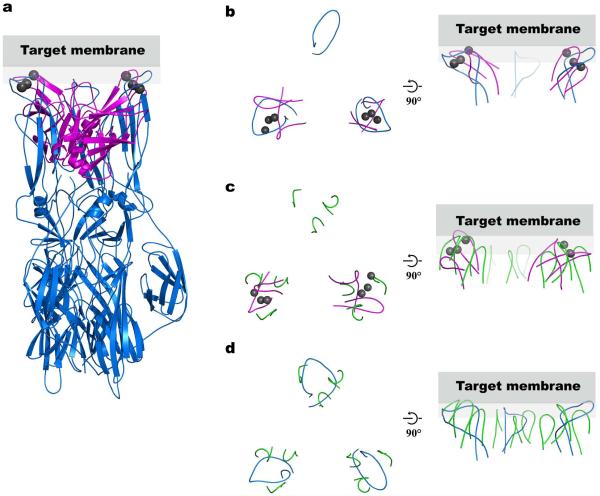
Figure 2.
Comparison of the structural arrangement of the Syt3 Ca2+-binding loops with Semliki Forest virus E1 fusion loops and Rhesus Rotavirus VP5 membrane-interacting loops. (a) We superimposed two of the three E1 loops of the Semliki Forest virus E1 protein (blue, PDB ID 1RER21) on the Ca2+-binding loops of Syt3 (magenta, Ca2+ ions are in gray). (b–d) A top view looking from the target membrane onto the proteins is shown in the left panels, and a side view in the right panels. Gray rectangles represent the target membrane. Only the fusion membrane-interacting loops are shown in all panels for clarity. (b) Top view of panel (a) showing only the superimposed loops. (c) We superimposed two of the three membrane-interacting loop regions of the Rhesus Rotavirus VP5 protein trimer (green, PDB ID 1SLQ22) on the Ca2+-binding loops of Syt3 (magenta, Ca2+ ions are in gray). (d) Superposition of the Semliki Forest virus E1 protein (blue) and the membrane-interacting loops of Rhesus Rotavirus VP5 protein (green) using all three loop regions. Both E1 of Semliki Forest virus and VP5 of Rhesus Rotavirus are trimers in their post-fusion state. In addition to structural similarities of the Semliki Forest virus fusion loops and the membrane interacting loops of VP5 Rhesus Rotavirus, these loops also share sequence homology22. The fusion/cell entry mechanism of non-enveloped rotavirus is not fully understood, however it is able to mediate cell-cell fusion in vitro47.