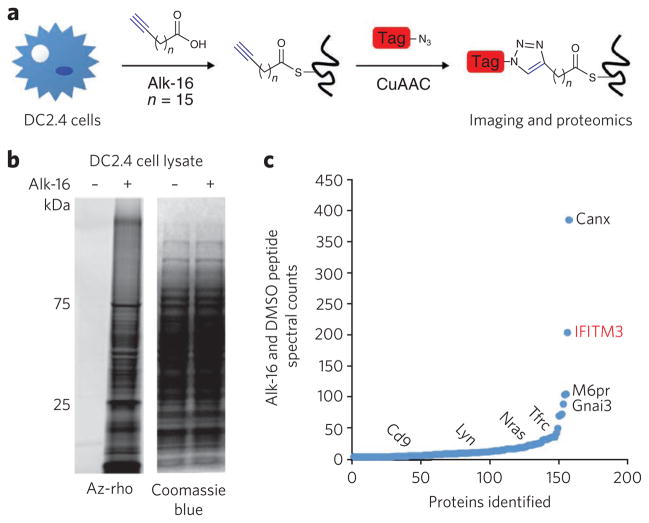
Figure 1. Visualization and identification of palmitoylated proteins in DC2.4 cells.
(a) Metabolic labeling of cells with alk-16 palmitate reporter and subsequent CuAAC ligation with bioorthogonal detection tags for imaging or proteomics. (b,c) DC2.4 cells were incubated for 2 h with 50 mM alk-16 or DMSO as a control. In b, cell lysates were reacted with az-rho by CuACC, and proteins were separated by SDS-PAGE for visualization by fluorescence gel scanning. Coomassie blue staining demonstrates comparable loading. In c, cell lysates were reacted with az-diazo-biotin by CuAAC for enrichment of alk-16–labeled proteins with streptavidin beads and identification by mass spectrometry. For each identified protein, the ratio of peptide spectral counts from the alk-16 and DMSO samples was plotted. Several known palmitoylated proteins are shown in black. IFITM3, the highest-ranked candidate palmitoylated protein, is shown in red.