Abstract
BRCA1 and BRCA2 screening in women at high-risk of breast cancer results in the identification of both unambiguously defined deleterious mutations and sequence variants of unknown clinical significance (VUS). We examined a population-based sample of young women with contralateral breast cancer (CBC, n=705) or unilateral breast cancer (UBC, n=1398). We identified 470 unique sequence variants, of which 113 were deleterious mutations. The remaining 357 VUS comprised 185 unique missense changes, 60% were observed only once, while 3% occurred with a frequency of >10%. Deleterious mutations occurred three times more often in women with CBC (15.3%) than in women with UBC (5.2%), whereas combined, VUS were observed in similar frequencies in women with CBC and UBC. A protein alignment algorithm defined 16 rare VUS, occurring at highly conserved residues and/or conferring a considerable biochemical difference, the majority located in the BRCA2 DNA-binding domain. We confirm a multiplicity of BRCA1 and BRCA2 VUS that occur at a wide range of allele frequencies. Although some VUS inflict chemical differences at conserved residues, suggesting a deleterious effect, the majority are not associated with an increased risk of CBC.
Keywords: Breast cancer, BRCA1, BRCA2, contralateral, risk, deleterious mutation, unclassified variants
INTRODUCTION
Women with a BRCA1 (MIM# 113705) or BRCA2 (MIM# 600185) germline mutation are at high risk of developing breast and ovarian cancer (Stratton and Rahman, 2008, Begg et al., 2008). Mutation screening is widely available from health care, research, and commercial laboratories. Thousands of different disease-associated mutations have been identified (Breast Cancer Information Core, BIC). Most deleterious mutations introduce premature termination codons through small frameshift deletions or insertions, nonsense or splice junction alterations, or large deletions or duplications. Some splice site mutations and large rearrangements do not change the reading frame, but result in a loss or gain of one to several exons, which is thought to compromise the gene function. Deleterious missense mutations are typically confined within specific residues of functional motifs. However, the risk contribution of numerous other sequence variants remains unclear. These ‘variants of unknown significance’ (VUS) include missense changes and small in-frame deletions and insertions, coding synonymous nucleotide substitutions that do not lead to amino acid shifts, as well as alterations in non-coding intervening sequences (IVS) or in untranslated exonic regions (UTRs).
Classifying VUS remains a great challenge for designing tailored genetic counseling and disease prevention strategies. First, VUS greatly outnumber known deleterious mutations and include both unidentified deleterious mutations as well as neutral variants with no clinical importance. As individual VUS are rare, the usual genetic approach of linkage/segregation or association analysis typically used to classify sequence variants is difficult to apply. As homozygosity or compound heterozygosity for deleterious mutations are embryonically lethal or associated with severe syndromes, VUS that co-occur with a known disease-associated mutation in trans can be classified as not being high-risk alleles. However, this approach relies on the correct determination of allelic phases (Judkins et al., 2005; Tavtigian et al., 2006). A variety of functional assays have been developed for BRCA1 including transcription activation small colony phenotype rescue of radiation resistance and ubiquitin ligase activity; and for BRCA2, assays for homologous recombination, crosslink dependent survival, centrosome amplification or rescue of mouse embryonic stem cell viability (Carvalho et al., 2007; Kuznetsov et al., 2008). Although potentially excellent strategies, they are mostly restricted to variants of certain domains and require laborious optimization. In the absence of a universal functional assay, in silico predictions of missense variants by assessment of phylogenetic conservation and severity in modification of biophysical characteristics of amino acids have also been utilized (Abkevich et al., 2004; Tavtigian et al., 2006). Moreover, all coding sequence alterations, including synonymous variants, can be assessed for potential to disrupt exonic splicing enhancer and silencer elements, or introduce cryptic splice sites, and should be considered along with variants in the exon/intron splice junction consensus sites as potentially affecting transcript processing. Multifactorial likelihood-ratio models have been developed to integrate information from these various sources, including tumor histopathological characteristics, providing likelihood ratios for or against disease causality for a considerable number of VUS (Goldgar et al., 2004; Chenevix-Trench et al., 2006; Easton et al., 2007). Nevertheless, the majority of variants studied remain of unknown clinical importance.
The Women’s Environment, Cancer, and Radiation Epidemiology (WECARE) Study was established to examine the combined roles of postoperative radiation exposure and genetic susceptibility in the etiology of contralateral breast cancer (CBC). A large multi-center, population-based series of asynchronous CBC and a reference group of matched unilateral breast cancer (UBC) was designed (Bernstein et al., 2004). We previously reported results from this series regarding estimates of risk variation in carriers of deleterious BRCA gene mutations (Begg et al., 2008). In the present study, we report results on the frequency and pattern of all types of BRCA1 and BRCA2 sequence variants. We used in silico analysis to predict the functional consequence of missense variants and CBC-UBC comparisons of groupings of variants to explore their association with breast cancer risk.
MATERIALS AND METHODS
Study population
The WECARE Study population included 705 women with CBC and 1398 women with UBC who were ascertained through five population-based cancer registries. Controls (women with UBC) were individually matched to the cases (women with CBC) on age, date of diagnosis, race, and registry (Bernstein et al., 2004). All were diagnosed with their first primary breast cancer at age 54 years or younger (mean age 46 years) and the mean interval from first to second breast cancer in the CBC group was 5 years (range 1–16 years). UBC controls were also counter-matched on radiation treatment (Bernstein et al., 2004). The proportion of women of Jewish ancestry was similar by CBC-UBC status (7.7%). A first-degree (affected mother, sister or daughter) family history of breast cancer was reported more frequently in women with CBC (32%) than in women with UBC (21%).
BRCA gene analysis and quality controls
The complete coding sequences of BRCA1 and BRCA2 were screened for variations by denaturing high-performance liquid chromatography (DHPLC) and sequence analysis, including a stepwise laboratory quality control (QC) scheme as previously described (Begg et al., 2008). In total, 139 samples were tested in the inter-lab QC step. Of the 9,869 fragments tested, discrepant results were observed for 52 (0.53%) of the fragments. For the intra-lab QC, of the 21,158 fragments tested, from 298 samples, discrepant results were observed for 111 (0.52%) fragments. In terms of the consistency of classifying deleterious mutation carrier status, among the 298 women whose specimens were tested twice within the same screening laboratory, only two (0.67%) had discrepant results, and among the 139 women whose samples were re-screened by the centralized laboratory, three (2.16%) had discrepant results. When QC results were un-blinded, discrepant results were resolved.
Sequence variation nomenclature
All sequence variants were named and are referred to in the text according to the nomenclature used by Human Genome Variation Society (HGVS) (http://www.hgvs.org) recommendation guidelines, using the A of the ATG-translation initiation codon as nucleotide +1 (den Dunnen and Antonarakis, 2000). Mutations are also provided using the BIC (http://research.nhgri.nih.gov/bic) nomenclature, with nucleotide numbering starting at the first transcribed base of BRCA1 (GenBank U14680.1) and BRCA2 (NM_000059.1). In Supp. Tables S1 and S2, all sequence variants are listed according to both BIC and HGVS.
In silico analyses
BRCA missense variants were analyzed using web-based algorithms, using information that is entirely external to the WECARE Study data. Align-GVGD (http://agvgd.iarc.fr/) combines the biophysical characteristics (side-chain composition, polarity and volume) of amino acids and protein multiple sequence alignments (Grantham Variation (GV) and Grantham Deviation (GD) scores) to predict where amino acid substitutions fall in a spectrum from enriched deleterious to enriched neutral, providing GV and GD scores (0 to >200) and graded classifiers (C0 to C65) (Tavtigian et al., 2006). Alignments were done with up to 13 BRCA1 sequences and 12 BRCA2 sequences to the depth of Xenopus laevis (frog), Tetraodon nigroviridis (pufferfish - green spotted) or Strongylocentrotus purpuratus (purple sea urchin).
Definition of deleterious mutations
Sequence variants were categorized based on their predicted effect on the mRNA and amino acid level and defined as deleterious mutations according to the following established (BIC) criteria: (1) all frameshift and nonsense variants with the exception of the neutral stop codon BRCA2 c.9976A>T (BIC: K3326X) (Mazoyer et al., 1996), and other variants located 3′ thereof; (2) all IVS variants occurring in the consensus splice acceptor or donor sequence sites, either within 2 bp of exon-intron junctions or when experimentally demonstrated to result in abnormal mRNA transcript processing; (3) missense variants that have been conclusively demonstrated, on the basis of data from linkage analysis of high risk families, functional assays or biochemical evidence, to have a deleterious effect on known functional regions. In this study, all variants not previously identified as being deleterious by these criteria were considered “VUS”.
Statistical Analyses
Age-adjusted conditional logistic regression was used to estimate the relative risks (RR) and corresponding 95% confidence intervals (95% CI) of common VUS as well as of rare variants grouped by type (deleterious, missense VUS, synonymous VUS, IVS VUS). Models included an “offset” term to account for the counter-matching. All analyses were conducted using SAS TPHREG (SAS Institute, Cary, NC).
RESULTS
Deleterious mutations identified
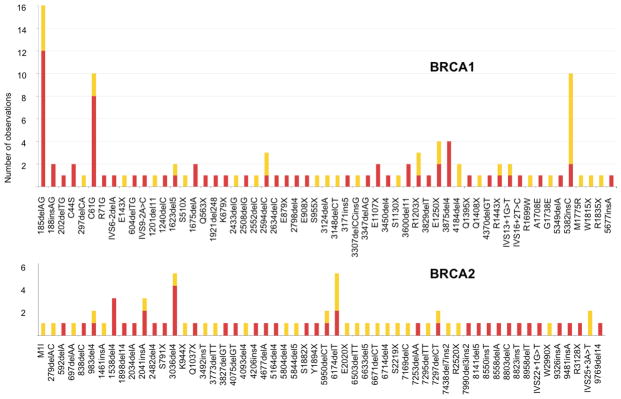
The mutation screening of 2103 participants (705 CBC and 1398 UBC) resulted in the discovery of 113 unique deleterious mutations, 57 in BRCA1 and 56 in BRCA2 (Figure 1, Supp. Table S1). These variants, and the corresponding familial aggregations of breast cancer in their relatives, were used in an earlier article to calculate penetrance estimators (Begg et al., 2008). The variants included 73 small frameshift deletions or insertions, 26 nonsense mutations, 7 missense and 7 splice site mutations. Five of the 7 deleterious splice site mutations were located within 2 bp from the exon/intron boundaries. BRCA2 c.9501+3A>T (BIC: IVS25+3A>T) was classified as deleterious because segregation/clinical data are supportive and cDNA analysis confirms aberrant transcript splicing, i.e. exon 25 skipping, resulting in a frameshift and an almost immediate premature stop codon. BRCA1 c.211A>G (BIC: R71G) is located in exon 5, two nucleotides from the 3′-exon/intron junction, and shown to introduce an aberrant cryptic splice site (Vega et al., 2001). All 7 splice site mutations were predicted to give rise to out-of-frame transcripts leading to premature stop codons. The 7 missense mutations defined as deleterious included BRCA1 c.130T>A (BIC: C44S) and c.181T>G (BIC: C61G), both occurring at zinc-ligating residues in the RING domain (Morris et al., 2006), and repeatedly found to segregate with disease in breast-ovarian cancer families in Scandinavia or globally. BRCA1 c.5095C>T (BIC: R1699W), c.5123C>A (BIC: A1708E), c.5213G>A (BIC: G1738E) and c.5324T>G (BIC: M1775R) all affect structurally and functionally critical residues in the BRCT domains and are mutations described previously in breast cancer families (Vallon-Christersson et al., 2001; Olopade et al., 2003; Williams et al., 2003). BRCA2 c.3G>A (BIC: M1I) disrupts the translation initiation codon and was classified as deleterious since a possible re-initiation at Met-124 would result in an amino-terminal truncated protein lacking important regulatory regions (Xia et al., 2006). Finally, three sequence variants – BRCA2 c.9976A>T (BIC: K3326X), c.10095delCins11 (BIC: 10323delCins11) and c.10150C>T (BIC: R3384X) – predicted to result in protein truncation were ruled as exceptions that could not be classified because of their location near the 3′-end and possibly dispensable part of the gene. Thus, the most carboxy-terminal deleterious mutation in BRCA2 was c.9541_9554del14 (BIC: 9769del14) and c.5558dupA (BIC: 5677insA) in BRCA1, the latter predicted to result in a protein truncated by only the last 11 amino acids. Our approach to classifying mutations as deleterious follows the approach currently used in oncogenetic clinics and is compatible with classifications in the BIC. Our screening method did not target larger genomic deletions or duplications. Thus, it is possible that some deleterious mutations may have been missed.
Figure 1.
Spectrum of deleterious BRCA1 (n=57) and BRCA2 (n=56) mutations detected in 705 women with CBC (red) and 1398 women with UBC (yellow), displayed by gene 5′ to 3′ location and number of observations. Mutation nomenclature according to BIC.
Frequency of deleterious mutations in women with CBC and UBC
A total of 181 (8.6%) of 2103 participants were found to carry one of the 113 unique deleterious BRCA mutations (Figure 1, Supp. Table S1). No woman carried more than one deleterious mutation. Mutations were more common in BRCA1 (n=109) than in BRCA2 (n=72). Multiple women carried known founder mutations such as BRCA1 c.68_69delAG (BIC: 185delAG) (n=16), c.181T>G (BIC: C61G) (n=10), c.5266dupC (BIC: 5382insC) (n=10), and BRCA2 c.5946delT (BIC: 6174delT) (n=5) and c.2808_2811delACAA (BIC: 3036del4) (n=5). Eighty-nine of the 113 mutations were detected only once (Figure 1). Deleterious mutations were nearly three times more common in women with CBC (15.3%) than in women with UBC (5.2%). Carrying a mutation in either BRCA1 or BRCA2 was associated with a 4-fold increased risk of CBC (95% CI 2.8–5.7) as compared to not having a deleterious mutation (Table 1). BRCA1 mutation carriers had a 4.5-fold increased risk (95% CI 2.8–7.1) and carriers of a BRCA2 mutation had a 3.4-fold increased risk (95% CI 2.0–5.8) of CBC. The increased risk of CBC was evident irrespective of mutation type, i.e. frameshift, nonsense, splice site or missense mutation (Table 1) (Malone et al., submitted 2009).
Table 1.
Risk of CBC compared to UBC, in relation to known deleterious mutations in BRCA1 and BRCA2.
| Carrier Status | CBCa | UBCa | Adjb % | RRc | 95% CI | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| No known deleterious mutation in BRCA1/2 d | 597 | 84.7 | 1325 | 94.8 | 95.1 | 1.0 | Ref. |
| Deleterious mutation in BRCA1 or BRCA2 | 108 | 15.3 | 73 | 5.2 | 4.9 | 4.0 | 2.8–5.7 |
| Deleterious mutation in BRCA1 | 67 | 9.5 | 42 | 3.0 | 2.6 | 4.5 | 2.8–7.1 |
| Deleterious mutation in BRCA2 | 41 | 5.8 | 31 | 2.2 | 2.4 | 3.4 | 2.0–5.8 |
| Frameshift, nonsense, splice mutation in BRCA1 | 56 | 7.9 | 37 | 2.6 | 2.4 | 4.2 | 2.5–7.0 |
| Deleterious missense mutation in BRCA1 | 11 | 1.6 | 5 | 0.4 | 0.2 | 6.3 | 2.0–19.9 |
| Frameshift, nonsense, splice mutation in BRCA2 | 41 | 5.8 | 30 | 2.1 | 2.3 | 3.6 | 2.1–6.1 |
| Deleterious missense mutation in BRCA2 | 0 | 0 | 1 | 0.1 | 0.1 | - | - |
| BRCA1, 3′end mutations e | 3 | 0.4 | 10 | 0.7 | 0.5 | 0.7 | 0.2–3.2 |
| BRCA1, all other truncating mutations | 53 | 7.5 | 27 | 1.9 | 1.8 | 5.3 | 3.0–9.3 |
| BRCA2, OCCR mutations f | 11 | 1.6 | 11 | 1.1 | 1.1 | 3.2 | 1.3–8.0 |
| BRCA2, all other deleterious truncating mutation | 30 | 4.3 | 19 | 1.1 | 1.2 | 3.8 | 1.9–7.3 |
Women with UBC (N=1398) served as reference group for women with CBC (N=705).
UBC proportions computed as if UBCs were randomly selected.
Relative risk was adjusted for counter-matching and age at first primary; 95%CI: 95% confidence interval.
Wildtype and all variants of unknown significance that have not earlier been classified as deleterious (see Supp. Table S2).
5382insC, W1815X, R1835X and 5677insA of BRCA1 (GenBank U14680.1), nomenclature according to BIC.
OCCR (Ovarian Cancer Cluster Region) premature stop codon occurring within nt. 3059–6629 of BRCA2 (GenBank NM_000059.1). Mutation nomenclature according to HGVS is found in Supp. Table S1.
Effect of position of deleterious mutations
A lower frequency of CBC patients with mutations located in the 3′ part of BRCA1 as compared with the 5′ and middle-region of BRCA1 was noted (Figure 1). The RR of CBC in the 13 women carrying the most 3′ located truncating mutations (here including c.5266dupC (BIC: 5382insC), c.5444G>A (BIC: W1815X), c.5503C>T (BIC: R1835X) and c.5558dupA (BIC: 5677insA)) was 0.7, while the RR was 5.3 in the 80 women carrying any other truncating BRCA1 mutation (Table 1). No such effect for corresponding 3′ located truncating BRCA2 mutations remained after excluding c.9976A>T (BIC: K3326X), c.10095delCins11 (BIC: 10323delCins11) and c.10150C>T (BIC: R3384X). There was no significant difference regarding the risk of CBC in women with premature stop codons occurring within or outside (either N- or C-terminal) the ‘ovarian cancer-cluster region’ (OCCR, nucleotides 3059–6629; GenBank NM_000059.1) of BRCA2 (Table 1) (Gayther et al., 1997).
Classification of VUS
Screening of 2103 women resulted in the identification of an additional 357 unique sequence variants that were not classified as deleterious (complete list in Supp. Table S2). These 357 variants, including both VUS and suspected (BIC) neutral variants, were broadly categorized according to type into: non-synonymous (n=185), synonymous (n=69), and IVS alterations (n=91). The results from these 345 variants are summarized in Tables 2a and 2b. In addition we found 4 rare in-frame deletions, 5 variants in UTRs, 1 neutral frameshift (BRCA2 c.10095delCins11 (BIC: 10323delCins11)) and 2 neutral nonsense (BRCA2 c.9976A>T (BIC: K3326X) and c.10150C>T (BIC: R3384X)) variants. The majority (n=210) of the 357 variants were observed only once while 81 other variants had a minor allele frequency (MAF) less than 0.1% (observed 4 times or less among the 4206 alleles). In contrast there were 16 common variants with MAF>10%. A substantial number (n=133) of variants found once or a few times have not been reported earlier (in BIC) (highlighted in blue in Supp. Table S2). This observation should, however, be interpreted cautiously since VUS are probably less frequently reported and are likely under-represented in public databases. Interestingly, the numbers of coding variants found in the BRCA genes are high considering that the average gene contains only a handful (~4) of coding SNPs with an allele frequency of at least a few percent in the population. Typically, non-synonymous SNPs not only occur less often but also have lower MAF, often less than 5% (Cargill et al., 1999).
Table 2a.
MAF of BRCA1 sequence VUSa, identified in women with CBC or UBC.
| Typeb | MAF | CBC | UBC | Relative Risk (95% CI) c,d |
|---|---|---|---|---|
| Missense | ||||
| 53 unique variants | <0.1% | 21 | 46 | 1.0 (0.6–1.7) |
| 4 unique variants | 0.1–0.5% | 9 | 35 | 0.6 (0.3–1.2) |
| 2 unique variants | 0.5–2.5% | 42 | 81 | 1.1 (0.8–1.7) |
| Q356R | 6.0% | 65 | 165 | 0.8 (0.6–1.2) |
| D693N | 7.1% | 87 | 180 | 1.1 (0.8–1.4) |
| E1038G | 25% | 271 | 646 | 0.9 (0.7–1.0) |
| K1183R | 26% | 272 | 642 | 0.9 (0.7–1.1) |
| P871L | 26% | 278 | 668 | 0.8 (0.7–1.0) |
| S1613G | 29% | 291 | 693 | 0.8 (0.7–1.0) |
| Synonymous | ||||
| 13 unique variants | <0.1% | 4 | 15 | 0.6 (0.2–1.7) |
| 2 unique variants | 0.1–0.5% | 2 | 9 | 0.5 (0.1–2.2) |
| no variant | 0.5–2.5% | - | - | - |
| L771L | 25% | 268 | 640 | 0.8 (0.7–1.0) |
| S1436S | 26% | 269 | 653 | 0.8 (0.7–1.0) |
| S694S | 28% | 278 | 676 | 0.8 (0.7–1.0) |
| IVS | ||||
| 32 unique variants | <0.1% | 11 | 21 | 1.4 (0.6–3.0) |
| 4 unique variants | 0.1–0.5% | 10 | 19 | 1.2 (0.6–2.7) |
| no variant | 0.5–2.5% | - | - | - |
| IVS17−53C>T | 2.7% | 24 | 77 | 0.7 (0.4–1.1) |
| IVS7−34C>T | 19% | 222 | 495 | 1.0 (0.8–1.2) |
| IVS8−58delT | 28% | 285 | 672 | 0.9 (0.7–1.1) |
| IVS7+36del14 | 33% | 314 | 745 | 0.8 (0.7–1.0) |
Individuals with rare variants are aggregated within MAF categories. Common variants (MAF>2.5%) are listed separately. Frequencies correspond to combined frequencies of heterozygotes or homozygote variants.
In addition to the variant types listed, 2 women with CBC and 7 women with UBC had rare variants that were either in-frame deletions or variants in the untranslated regions. BRCA1 (GenBank U14680.1) mutation nomenclature according to BIC. IVS=intervening sequence (noncoding). Mutation nomenclature according to HGVS is found in Supp. Table S1.
95% confidence interval.
The reference group for each RR consists of women carrying all other types of VUSs and excludes carriers of known deleterious BRCA mutations.
Table 2b.
MAF of BRCA2 sequence VUSa, identified in women with CBC or UBC.
| Typeb | MAF | CBC | UBC | Relative Risk (95%CI) c,d |
|---|---|---|---|---|
| Missense | ||||
| 102 unique variants | <0.1% | 46 | 72 | 1.4 (0.9–2.0) |
| 10 unique variants | 0.1–0.5% | 24 | 57 | 0.9 (0.5–1.5) |
| 4 unique variants | 0.5–2.5% | 22 | 64 | 0.7 (0.4–1.2) |
| T1915M | 2.6% | 26 | 74 | 0.8 (0.5–1.2) |
| N289H | 3.5% | 44 | 92 | 1.1 (0.7–1.6) |
| N991D | 3.5% | 43 | 92 | 1.0 (0.7–1.5) |
| N372H | 28% | 310 | 627 | 1.2 (1.0–1.4) |
| Synonymous | ||||
| 40 unique variants | <0.1% | 18 | 34 | 1.2 (0.7–2.2) |
| 6 unique variants | 0.1–0.5% | 19 | 37 | 1.1 (0.6–2.0) |
| no variant | 0.5–2.5% | - | - | - |
| H743H | 3.4% | 42 | 90 | 1.0 (0.7–1.5) |
| S455S | 3.5% | 44 | 89 | 1.1 (0.7–1.6) |
| V1269V | 16% | 177 | 419 | 0.9 (0.7–1.1) |
| S2414S | 19% | 207 | 477 | 1.0 (0.8–1.2) |
| K1132K | 23% | 267 | 581 | 1.1 (0.9–1.3) |
| IVS | ||||
| 41 unique variants | <0.1% | 20 | 33 | 1.4 (0.8–2.5) |
| 8 unique variants | 0.1–0.5% | 20 | 48 | 0.9 (0.5–1.6) |
| 1 unique variants | 0.5–2.5% | 8 | 23 | 0.7 (0.3–1.6) |
| IVS16−14T>C | 28% | 325 | 689 | 1.1 (0.9–1.3) |
Individuals with rare variants are aggregated within MAF categories. Common variants (MAF>2.5%) are listed separately. Frequencies correspond to combined frequencies of heterozygotes or homozygote variants.
In addition to the variants listed, 3 women with CBC and 3 women with UBC had rare variants that were either in-frame deletions, frameshifts, nonsense mutations or variants in the untranslated regions (UTR). In addition there is a 5′ UTR polymorphism (203G>A) that was observed in 269 CBC and 528 UBC (OR=1.0). BRCA2 (GenBank NM_000059.1) mutation nomenclature according to BIC. IVS=intervening sequence (noncoding). Mutation nomenclature according to HGVS is found in Supp. Table S1.
95% confidence interval.
The reference group for each RR consists of women carrying all other types of VUSs and excludes carriers of known deleterious BRCA mutations.
Frequency of VUS in women with CBC and UBC
Combined, rare VUS occurred in similar proportions in women with CBC and UBC (Tables 2a and 2b). Broadly, VUS, regardless of their MAFs’, occurred in similar frequencies in women with CBC and UBC for both BRCA1 variants (Table 2a) and BRCA2 variants (Table 2b). In these tables, rare variants have been grouped according to both type and MAF, while more common variants (MAF>2.5%) are listed individually. None of the sub-groups demonstrate a statistically significant effect on risk of CBC. Since these comparisons aggregate many individual variants we cannot on this basis rule out the possibility that there are a few rare variants that are actually associated with risk.
BRCA1 has four very common missense variants (c.2612C>T (BIC: P871L), c.3113A>G (BIC: E1038G), c.3548A>G (BIC: K1183R), c.4837A>G (BIC: S1613G)) with MAF>20%, and another two (c.1067A>G (BIC: Q356R and c.2077G>A (BIC: D693N)) with MAFs of 6% and 7% respectively. In BRCA2, one missense variant (c.1114A>C (BIC: N372H)) has a MAF>20% and three others (c.865A>C (BIC: N289H), c.2971A>G (BIC: N991D) and c.5744C>T (BIC: T1915M)) have MAFs of 2–4%. It is expected that these 10 are neutral variants of negligible functional and clinical significance. The observed relative frequencies of these variants in CBC versus UBC are consistent with the hypothesis that these variants do not affect the risk of breast cancer. However, homozygous status for BRCA2 His372 has previously been associated with a slight increased risk of breast cancer (Healey et al., 2000). We saw no statistically significant increased risk of CBC in homozygous or heterozygous His-372 carriers as compared to WT (RR for heterozygous 1.19 95%CI: 0.97–1.47, and RR for homozygous 1.12 95%CI: 0.77–1.63).
Functional assessment of non-synonymous VUS
We used Align-GVGD to further assess the functional effect of missense variants, with alignment to 13 BRCA1 and 12 BRCA2 ortholog sequences down to sea urchin. Six VUS (BRCA1 c.2596C>T (BIC: R866C), BRCA2 c.4585G>A (BIC: G1529R), c.7878G>C (BIC: W2626C), c.7988A>T (BIC: E2663V), c.7994A>G (BIC: D2665G) and c.9154C>T (BIC: R3052W)) occurred at strongly conserved residues (GV=0) and had a GD≥65. Thus, these were inferred to belong to the class (C65) of substitutions most likely to interfere with function and were in this respect comparable to five of the missense mutations a priori classified as deleterious (the remaining, BRCA1 c.5324T>G (BIC: M1775R) had a GV=14.30 and was inferred to class C45, while BRCA2 c.3G>A (BIC: M1I) had a GV=0 but GD as low as 10.12). Two other BRCA2 variants (c.9104A>G (BIC: Y3035C) and c.10045A>G (BIC: T3349A)) were defined as interfering with function (A-GVGD class C55), an additional 8 VUS had either a low GV or high GD score which lifted them above class C0 and the remaining 169 amino acid substitutions were less likely to compromise function. Details of 16 variants are presented in Table 3 with corresponding details of all VUS missense variants in Supp. Table S3. Lowering the sequence alignment stringency to the level of pufferfish and frog, respectively, led to another 7 and 16 VUS predicted to be functionally defective (C15-C55, Supp. Table S3).
Table 3.
(a) BRCA1 and BRCA2 VUS identified in CBC and UBC as potentially deleterious by A-GVGD protein alignment algorithm (to the depth of Sea Urchin), yielding a GV=0 and/or a GD≥65. (b) A-GVGD scores, CBC and UBC numbers of BRCA1 and BRCA2 missense variants earlier classified as being deleterious mutations.
| (a) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| A-GVGD | |||||||||
| Gene | Exon | NTa | Base Ch | BICb | Class | GV | GD | CBC | UBC |
| BRCA1 | 7 | 433 | A>G | Y105C c,d | C15 | 120 | 131 | 0 | 1 |
| BRCA1 | 8 | 655 | A>G | Y179C | C35 | 40 | 182 | 0 | 2 |
| BRCA1 | 11 | 2640 | C>T | R841W | C15 | 111 | 78 | 2 | 6 |
| BRCA1 | 11 | 2715 | C>T | R866C c,d | C65 | 0 | 180 | 0 | 1 |
| BRCA1 | 12 | 4291 | T>C | I1391T | C25 | 30 | 70 | 1 | 0 |
| BRCA1 | 22 | 5520 | G>T | G1801C | C15 | 157 | 107 | 0 | 1 |
| BRCA2 | 11 | 4813 | G>A | G1529R c,d | C65 | 0 | 125 | 2 | 0 |
| BRCA2 | 17 | 8106 | G>C | W2626C | C65 | 0 | 214 | 1 | 0 |
| BRCA2 | 17 | 8107 | A>T | I2627F | C15 | 0 | 21 | 0 | 1 |
| BRCA2 | 18 | 8216 | A>T | E2663V | C65 | 0 | 121 | 0 | 1 |
| BRCA2 | 18 | 8222 | A>G | D2665G c,d | C65 | 0 | 94 | 1 | 1 |
| BRCA2 | 18 | 8267 | A>G | D2680G | C15 | 44 | 75 | 0 | 1 |
| BRCA2 | 22 | 9078 | G>T | K2950N d | C35 | 26 | 79 | 3 | 0 |
| BRCA2 | 23 | 9332 | A>G | Y3035C | C55 | 21 | 192 | 1 | 1 |
| BRCA2 | 24 | 9382 | C>T | R3052W | C65 | 0 | 101 | 0 | 1 |
| BRCA2 | 27 | 10273 | A>G | T3349A c,d | C55 | 0 | 58 | 0 | 1 |
| Total | 11 | 18 | |||||||
| (b) | |||||||||
| BRCA1 | 3 | 249 | T>A | C44S e | C65 | 0 | 112 | 2 | 0 |
| BRCA1 | 5 | 300 | T>G | C61G e | C65 | 0 | 158 | 8 | 2 |
| BRCA1 | 18 | 5214 | C>T | R1699W e | C65 | 0 | 101 | 0 | 1 |
| BRCA1 | 18 | 5242 | C>A | A1708E e | C65 | 0 | 107 | 1 | 0 |
| BRCA1 | 20 | 5332 | G>A | G1738E e | C65 | 0 | 98 | 0 | 1 |
| BRCA1 | 21 | 5443 | T>G | M1775R e | C45 | 14 | 91 | 0 | 1 |
| BRCA2 | 2 | 231 | G>A | M1I e | C0 | 0 | 10 | 0 | 1 |
| Total | 11 | 6 | |||||||
NT: nucleotide. BRCA1 (GenBank U14680.1), BRCA2 (GenBank NM_000059.1), according to BIC.
BIC: nomenclature according to Breast Cancer Information Core (see Supp. Table S2 for HGV nomenclature).
Classified as of no clinical interest in BIC.
Classified as neutral in the likelihood-ratio model developed by Easton et al. (2007).
BRCA1 and BRCA2 missense variants a priori classified as deleterious mutations in the present study.
All 8VUS with the highest likelihood of being deleterious (A-GVGD class C55-C65 in alignment down to sea urchin) were rare (found once or twice), while another 8VUS reaching class C15-C35 in alignment to sea urchin were observed a total of 18 times. However, the aggregated ratio of CBC (11) to UBC (18) only slightly exceeds the reference ratio of one CBC to every two UBCs in the study. The 15 VUS assigned to class C15-C65 in alignments to pufferfish or frog, but to C0 in alignment to sea urchin, were observed 78 times. The latter group included BRCA1 c.134A>C (BIC: K45T) in the RING domain (see below), as well as BRCA2 c.8851G>A (BIC: A2951T) (class C55), which was observed 22 times and had an elevated frequency in women with CBC (10 CBC versus 12 UBC). Interestingly, this variant has earlier been suggested as a moderate-risk predisposing allele (Hammet et al., 2008).
Functional domains affected by non-synonymous VUS
None of the missense or in frame deletion variants identified in the present study affected the numerous putative phosphorylation sites in BRCA1 or BRCA2, nor were they found in the nuclear localization signals of BRCA1 (amino acids 503–508 and 606–615) nor of BRCA2 (amino acids 3266–3269 and 3311–3315). Besides the two deleterious mutations at Cys44 and Cys61, a third variant (c.134A>C (BIC: K45T), observed once) affected the BRCA1 RING domain (amino acids 1–100). Although not as strongly conserved as the RING domain cysteine residues, Lys45 is part of a beta-sheet and K45T disrupts a salt bridge to Glu75, likely to weaken the folding and interaction with the E2 ubiquitin-conjugating enzyme UbcH5a (Morris et al., 2006). Several known deleterious missense mutations were found in the BRCA1 carboxy terminal region including the two BRCT domains (amino acids 1625–1825), but only one (c.5401G>T (BIC: G1801C)) of five other missense VUS in this region had an A-GVGD score suggesting a possible functional interference. The majority of BRCA2 VUS with an A-GVGD score suggesting a deleterious effect were located in the DNA binding domain (amino acids 2500–3098). Moreover, c.4585G>A (BIC: G1529R) affects a highly conserved residue in the Rad51 binding BRC4 motif of BRCA2 (Bignell et al., 1997). BRCA1 c.2596C>T (BIC: R866C) is located in a highly conserved region ‘motif 6’, still of unknown function, while BRCA1 c.4172T>C (BIC: I1391T) affects the coiled coil region (Abkevich et al., 2004).
DISCUSSION
Predisposition to breast cancer can be attributed to several levels of genetic susceptibility: rare high-risk alleles, rare moderate-risk alleles and common low-risk alleles (Stratton and Rahman, 2008). Deleterious mutations in BRCA1 and BRCA2 account for a considerable proportion of dominantly inherited breast cancer and have received wide acceptance in diagnostic testing and prevention. BRCA1 and BRCA2 mutation screening results in the identification of sequence variants that cannot be unequivocally referred to as deleterious or neutral. This is emphasized by the results from the present study of a high-risk population of young (<55 years) women with breast cancer. Using a carefully optimized DHPLC screening approach to analyze all coding exons and flanking intronic regions of BRCA1 and BRCA2, we identified 470 unique sequence variants in 2103 women. The majority (n=299) of variants were observed only once, however, there were some that were very common (MAF>10%). Based on documented knowledge on effects of variants that give rise to premature stop codons (via frameshift insertions or deletions, nonsense or consensus splice site sequence changes) or missense alterations at critical residues in functional domains, we defined 113 unique BRCA1 or BRCA2 variants as deleterious mutations. These were observed in 181 (8.6%) of the 2103 women. We confirmed earlier observations (BIC; Thomassen et al., 2008), of global or local founder effects for certain recurrent mutations and also describe a set of unique mutations as previously reported (Begg et al., 2008; Malone et al., submitted 2009). Deleterious mutations were three times as common in women with CBC (15.3%) than in women with UBC (5.2%), and carrying a deleterious mutation in either BRCA1 or BRCA2 conferred an approximately 4-fold increased risk of CBC among survivors of a first breast cancer (Begg et al., 2008; Malone et al., submitted 2009).
Previous studies have suggested a genotype-phenotype correlation within BRCA1 in which women with truncating mutations 3′ of the exon 12/13 boundary show a significantly decreased ovarian to breast cancer ratio (Gayther et al., 1995; Holt et al., 1996; Thompson et al., 2002). Our study design precluded assessment of the breast-ovarian cancer mutation location question but allowed us to assess whether the occurrence of CBC was related to location. We observed a lower frequency of CBC patients with mutations located in the 3′ part of BRCA1 as compared with the 5′ and middle-region of BRCA1. This was an unexpected finding considering that most transcripts with premature termination codons are targeted for degradation by the nonsense-mediated mRNA decay (NMD) surveillance pathway, resulting in no or low expression of nonsense transcripts and truncated protein products (Perrin-Vidoz et al., 2002; Conti and Izaurralde, 2005). However, transcripts bearing termination codons located <50 nucleotides from the last exon-exon junction (EEJ) or lacking a downstream EEJ (i.e., premature stop codons occurring in the last exon) may escape NMD. Thus, transcripts with premature stop codons occurring at BRCA1 codon 1807 or later may be translated and partly functional. Indeed, decreased but detectable expression of mutant protein has been found in HCC1937 cells carrying the BRCA1 5382insC mutation (Scully et al., 1999). However, despite the noted differences in the frequencies of women with CBC and UBC in the 5′ and 3′ parts of BRCA1, the numbers are small and results must be carefully interpreted. Truncating mutations in a central region of BRCA2 have been associated with an increased risk of developing ovarian cancer relative to breast cancer as compared to mutations occurring outside this OCCR bounds (Gayther et al., 1997; Thompson et al., 2001). It is less likely that escape from the NMD pathway explains this BRCA2 genotype-phenotype correlation (Ware et al., 2006), and we detected only a small, non-significant difference in the incidence of CBC in women with truncating mutations in a region overlapping with the OCCR. However, it cannot be excluded that expression of truncated BRCA proteins in variable degree interferes with normal BRCA function in tumorigenesis or retains some functions that influence tumor biology and DNA damage control in response to therapy in the absence of wildtype BRCA protein.
An even more complicated circumstance concerns the significance of expression of full-length BRCA proteins with missense mutations or small in-frame amino acid deletions/insertions. We identified seven missense variants that have been previously unambiguously defined as high-risk alleles due to known interference with function or translation initiation, or based on segregation analysis in affected families. These deleterious mutations were located at residues in the BRCA1 RING domain, important for ubiquitin-ligase activity, or in the BRCA1 BRCT-domains that interact with numerous proteins involved in transcription and DNA repair. However, we identified an additional 185 unique missense variants (and four in-frame single amino acid deletions) that could not straightforwardly be classified as risk-alleles. Again, the majority of these types of variants were rare and occurred in single individuals, while the minority were common. While it might be argued that the common variants are unlikely to be functional and should be referred to as neutral variants or polymorphisms, there is some suggestion that common BRCA missense variants may have a role as low-risk alleles (Healey et al., 2000; Hammet et al., 2008). We saw no evidence for increased risk associated with any of the common missense variants.
Theoretically, to further examine the role of each missense variant, one could take into account the multiplicity of functions and regions in both BRCA1 and BRCA2 that are involved in protein or DNA binding or constitute sites for enzymatic modification; however, the limited number of individual variants and lack of experimental validation would make such comparisons speculative and inconclusive. Therefore, we used publicly available protein alignment algorithms to more objectively evaluate the possible consequence of each amino acid substitution. Using Align-GVGD, we identified a set of 16 missense VUS (Table 3) that occurred at highly conserved (Grantham Variation=0) residues or conferred a considerable biochemical difference (Grantham Deviation≥65). Six of these occurred in BRCA1 and regions of unclear function such as the highly conserved motif 6 (Arg-866) or the coiled coil region (Ile-1391) (Abkevich et al., 2004), while ten were found in BRCA2. Interestingly, the majority (8/10) of the latter were located in the BRCA2 DNA binding domain (amino acids 2500–3098), an observation also made by others (Wu et al., 2005; Easton et al., 2007).
It is notable that the relative frequencies of CBC and UBC among the 16 VUS suggested by A-GVGD as candidate risk variants show no collective evidence of increased breast cancer risk (11/29 (38%) CBC, as compared with 705/1398 (34%) in the entire study). Furthermore, four (BRCA1 c.314A>G (BIC: Y105C), c.2596C>T (BIC: R866C), BRCA2 c.4585G>A (BIC: G1529R) and c.7994A>G (BIC: D2665G)) of the 16 VUS suggested by A-GVGD as candidate risk variants have been reported in BIC as of no clinical interest primarily based on co-occurrence (also in trans) with deleterious mutations and lack of segregation with disease in families, and six (BRCA1 c.314A>G (BIC: Y105C), c.2596C>T (BIC: R866C), BRCA2 c.4585G>A (BIC: G1529R), c.7994A>G (BIC: D2665G), c.8850G>T (BIC: K2950N) and c.10045A>G (BIC: T3349A)) were also classified as neutral in the likelihood-ratio model developed by Easton et al. (2007). However, two VUS scored as possibly deleterious were in agreement with Easton et al. (2007) (BRCA2 c.7878G>C (BIC: W2626C) and c.7988A>T (BIC: E2663V)) and two (BRCA2 c.7988A>T (BIC: E2663V) and c.9154C>T (BIC: R3052W)) were confirmed as deleterious in a recent study using a mouse embryonic stem cell assay (Kuznetsov et al., 2008). Thus, there is little evidence from this admittedly small list of candidates that tools based on evolutionary conservation are effective at identifying risk variants reliably.
In general, approximately two-thirds of randomly occurring point mutations in coding sequence would alter an amino acid, and the fact that non-synonymous coding SNPs comprise less than one half of coding SNPs in the genome implies a strong selection against amino acid altering changes (Cargill et al., 1999). A major interest in human genetics is to distinguish mutations that are functionally neutral from those that contribute to disease. Amino acid substitutions currently account for approximately half of the known gene lesions responsible for human inherited disease. It has been suggested that most rare missense variants are probably at least mildly deleterious and the accumulation of such low-risk variants may be the basis for complex diseases such as breast cancer (Kryukov et al., 2007; Stratton and Rahman, 2008). In the WECARE Study, we could only define a small fraction of all identified BRCA1 and BRCA2 missense variants as likely to have had a deleterious effect on breast cancer risk. The absence of an aggregate association between the remaining missense VUS and risk may indicate that there are very few additional clearly deleterious missense variants. However, this does not exclude the possibility that a few additional BRCA missense variants are functionally perturbed and associated with an increased breast cancer risk. For example, in this study we made no attempt to investigate the potential effect of sequence variants with respect to altering, adding or disrupting motifs of exonic splicing enhancers or silencers, or splice donor, acceptor or branch point sites. This analysis would include also synonymous and noncoding variants and add additional levels of complexity in interpreting the significance of BRCA sequence variants found in women with breast cancer.
In conclusion, this WECARE Study comprises a comprehensive effort in genetic screening of a large and uniform sample set, and resulted in a careful characterization of the landscape of BRCA1 and BRCA2 sequence variants in young women with breast cancer. One striking observation was the high frequency of rare missense variants that could not be unequivocally classified. We identified a number of new potentially deleterious missense mutations for further analysis and emphasize the large BRCA2 DNA binding domain as a possible target for additional candidates. Our overall conclusion, however, is that the majority of VUSs found in BRCA screening of affected women represent neutral alleles of no or little significance in the etiology of breast cancer.
Supplementary Material
Acknowledgments
Contract grant sponsor: National Cancer Institute, awards CA097397, CA131010 and CA098438.
WECARE STUDY COLLABORATIVE GROUP
Memorial Sloan Kettering Cancer Center (New York, NY): Jonine L. Bernstein Ph.D. (Study P.I.), Colin B. Begg. Ph.D., Marinela Capanu Ph.D., Xiaolin Liang M.D., Anne S. Reiner M.P.H., Tracy M. Layne M.P.H.; City of Hope (Duarte, CA): Leslie Bernstein Ph.D., Laura Donnelly-Allen; Danish Cancer Society (Copenhagen, Denmark): Jørgen H. Olsen M.D., D.M.Sc., Michael Andersson M.D., D.M.Sc., Lisbeth Bertelsen M.D., Ph.D., Per Guldberg Ph.D., Lene Mellemkjær Ph.D.;
Fred Hutchinson Cancer Research Center (Seattle, WA): Kathleen E. Malone Ph.D., Noemi Epstein; International Epidemiology Institute (Rockville, MD) and Vanderbilt University (Nashville, TN): John D. Boice Jr. Sc.D.; Lund University (Lund, Sweden): Åke Borg Ph.D., Therese Törngren M.Sc., Lina Tellhed, B.Sc.; National Cancer Institute (Bethesda, MD): Daniela Seminara Ph.D. M.P.H.; New York University (New York, NY): Roy E. Shore Ph.D., Dr.PH.; Norwegian Radium Hospital (Oslo, Norway): Anne-Lise Børresen-Dale Ph.D., Laila Jansen; University of California at Irvine (Irvine, CA): Hoda Anton-Culver, Ph.D., Joan Largent Ph.D. M.P.H.; University of Iowa (Iowa City, IA): Charles F. Lynch M.D., Ph.D., Jeanne DeWall M.A.; University of Southern California (Los Angeles, CA): Robert W. Haile DrPH., Bryan M. Langholz Ph.D., Duncan C. Thomas Ph.D., Shanyan Xue M.D., Nianmin Zhou, M.D, Anh T. Diep, Evgenia Ter-Karapetova; University of Southern Maine (Portland, ME):W. Douglas Thompson Ph.D.; University of Texas, M.D. Anderson Cancer Center (Houston, TX): Marilyn Stovall Ph.D., Susan Smith M.P.H.; University of Virginia (Charlottesville, VA): Patrick Concannon, Ph.D., Sharon Teraoka, Ph.D., Eric R. Olson, Nirasha Ramchurren, Ph.D.
References
- Abkevich V, Zharkikh A, Deffenbaugh AM, Frank D, Chen Y, Shattuck D, Skolnick MH, Gutin A, Tavtigian SV. Analysis of missense variation in human BRCA1 in the context of interspecific sequence variation. J Med Genet. 2004;41:492–507. doi: 10.1136/jmg.2003.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg CB, Haile RW, Borg A, Malone KE, Concannon P, Thomas DC, Langholz B, Bernstein L, Olsen JH, Lynch CF Anton-Culver H, Capanu M, Liang X, Hummer AJ, Sima C, Bernstein JL. Variation of breast cancer risk among BRCA1/2 carriers. JAMA. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JL, Langholz B, Haile RW, Bernstein L, Thomas DC, Stovall M, Malone KE, Lynch CF, Olsen JH, Anton-Culver H, Shore RE, Boice JD, Jr, Berkowitz GS, Gatti RA, Teitelbaum SL, Smith SA, Rosenstein BS, Børresen-Dale AL, Concannon P, Thompson WD WECARE study. Study design: evaluating gene-environment interactions in the etiology of breast cancer – WECARE Study. Breast Cancer Res. 2004;6:R199–214. doi: 10.1186/bcr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell G, Micklem G, Stratton MR, Ashworth A, Wooster R. The BRC repeats are conserved in mammalian BRCA2 proteins. Hum Mol Genet. 1997;6:53–58. doi: 10.1093/hmg/6.1.53. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Carvalho MA, Couch FJ, Monteiro AN. Functional assays for BRCA1 and BRCA2. Int J Biochem Cell Biol. 2007;39:298–310. doi: 10.1016/j.biocel.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevix-Trench G, Healey S, Lakhani S, Waring P, Cummings M, Brinkworth R, Deffenbaugh AM, Burbidge LA, Pruss D, Judkins T, Scholl T, Bekessy A, Marsh A, Lovelock P, Wong M, Tesoriero A, Renard H, Southey M, Hopper JL, Yannoukakos K, Brown M, Easton D, Tavtigian SV, Goldgar D, Spurdle AB kConFab Investigators. Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res. 2006;66:2019–2027. doi: 10.1158/0008-5472.CAN-05-3546. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, Tavtigian SV, Monteiro AN, Iversen ES, Couch FJ, Goldgar DE. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayther SA, Warren W, Mazoyer S, Russell PA, Harrington PA, Chiano M, Seal S, Hamoudi R, Van Rensburg EJ, Dunning AM, Love R, Evans G, Easton D, Clayton D, Stratton MR, Ponder BA. Germline mutations of the BRCA1 gene in breast and ovarian cancer families provide evidence for a genotype-phenotype correlation. Nat Genet. 1995;11:428–433. doi: 10.1038/ng1295-428. [DOI] [PubMed] [Google Scholar]
- Gayther SA, Mangion J, Russell P, Seal S, Barfoot R, Ponder BA, Stratton MR, Easton D. Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat Genet. 1997;15:103–115. doi: 10.1038/ng0197-103. [DOI] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Deffenbaugh AM, Monteiro AN, Tavtigian SV, Couch FJ. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet. 2004;75:535–544. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammet F, George J, Tesoriero AA, Jenkins MA, Schroen C, Smith L, Grabosch-Meehan A, Dite G, McCredie MR, Giles GG, Tavtigian SV, Hopper JL, Southey MC. Is BRCA2 c.9079 G > A a predisposing variant for early onset breast cancer? Breast Cancer Res Treat. 2008;109:177–179. doi: 10.1007/s10549-007-9624-6. [DOI] [PubMed] [Google Scholar]
- Healey CS, Dunning AM, Teare MD, Chase D, Parker L, Burn J, Chang-Claude J, Mannermaa A, Kataja V, Huntsman DG, Pharoah PD, Luben RN, Easton DF, Ponder BA. A common variant in BRCA2 is associated with both breast cancer risk and prenatal viability. Nat Genet. 2000;26:362–364. doi: 10.1038/81691. [DOI] [PubMed] [Google Scholar]
- Judkins T, Hendrickson BC, Deffenbaugh AM, Eliason K, Leclair B, Norton MJ, Ward BE, Pruss D, Scholl T. Application of embryonic lethal or other obvious phenotypes to characterize the clinical significance of genetic variants found in trans with known deleterious mutations. Cancer Res. 2005;65:10096–10103. doi: 10.1158/0008-5472.CAN-05-1241. [DOI] [PubMed] [Google Scholar]
- Holt JT, Thompson ME, Szabo C, Robinson-Benion C, Arteaga CL, King MC, Jensen RA. Growth retardation and tumour inhibition by BRCA1. Nat Genet. 1996;12:298–302. doi: 10.1038/ng0396-298. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SG, Liu P, Sharan SK. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat Med. 2008;14:875–81. doi: 10.1038/nm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone KE, Begg CB, Haile RW, Borg A, Concannon PJ, Tellhed L, Xue S, Teraoka S, Bernstein L, Capanu M, Reiner A, Riedel ER, Thomas DC, Mellemkjaer L, Lynch CF, Boice JD, Anton-Culver H, Bernstein JL. A population-based study of the relative and absolute risks of contralateral breast cancer associated with carrying a mutation in BRCA1 or BRCA2: Results from the WECARE Study. 2009 doi: 10.1200/JCO.2009.24.2495. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazoyer S, Dunning AM, Serova O, Dearden J, Puget N, Healey CS, Gayther SA, Mangion J, Stratton MR, Lynch HT, Goldgar DE, Ponder BA, Lenoir GM. A polymorphic stop codon in BRCA2. Nat Genet. 1996;14:253–254. doi: 10.1038/ng1196-253. [DOI] [PubMed] [Google Scholar]
- Morris JR, Pangon L, Boutell C, Katagiri T, Keep NH, Solomon E. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum Mol Genet. 2006;15:599–606. doi: 10.1093/hmg/ddi476. [DOI] [PubMed] [Google Scholar]
- Olopade OI, Fackenthal JD, Dunston G, Tainsky MA, Collins F, Whitfield-Broome C. Breast cancer genetics in African Americans. Cancer. 2003;97(Suppl):236–245. doi: 10.1002/cncr.11019. [DOI] [PubMed] [Google Scholar]
- Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11:2805–2814. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassen M, Hansen TV, Borg A, Lianee HT, Wikman F, Pedersen IS, Bisgaard ML, Nielsen FC, Kruse TA, Gerdes AM. BRCA1 and BRCA2 mutations in Danish families with hereditary breast and/or ovarian cancer. Acta Oncol. 2008;47:772–777. doi: 10.1080/02841860802004974. [DOI] [PubMed] [Google Scholar]
- Thompson D, Easton D Breast Cancer Linkage Consortium. Variation in cancer risks, by mutation position, in BRCA2 mutation carriers. Am J Hum Genet. 2001;68:410–419. doi: 10.1086/318181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Easton D. Variation in BRCA1 cancer risks by mutation position. Cancer Epidemiol Biomark Prev. 2002;11:329–336. [PubMed] [Google Scholar]
- Vallon-Christersson J, Cayanan C, Haraldsson K, Loman N, Bergthorsson JT, Brøndum-Nielsen K, Gerdes AM, Møller P, Kristoffersson U, Olsson H, Borg A, Monteiro AN. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet. 2001;10:353–360. doi: 10.1093/hmg/10.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega A, Campos B, Bressac-De-Paillerets B, Bond PM, Janin N, Douglas FS, Domenech M, Baena M, Pericay C, Alonso C, Carracedo A, Baiget M, Diez O. The R71G BRCA1 is a founder Spanish mutation and leads to aberrant splicing of the transcript. Hum Mutat. 2001;17:520–521. doi: 10.1002/humu.1136. [DOI] [PubMed] [Google Scholar]
- Ware MD, DeSilva D, Sinilnikova OM, Stoppa-Lyonnet D, Tavtigian SV, Mazoyer S. Does nonsense-mediated mRNA decay explain the ovarian cancer cluster region of the BRCA2 gene? Oncogene. 2006;25:323–328. doi: 10.1038/sj.onc.1209033. [DOI] [PubMed] [Google Scholar]
- Williams RS, Glover JN. Structural consequences of a cancer-causing BRCA1-BRCT missense mutation. J Biol Chem. 2003;278:2630–2635. doi: 10.1074/jbc.M210019200. [DOI] [PubMed] [Google Scholar]
- Wu K, Hinson SR, Ohashi A, Farrugia D, Wendt P, Tavtigian SV, Deffenbaugh A, Goldgar D, Couch FJ. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 2005;65:417–426. [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.