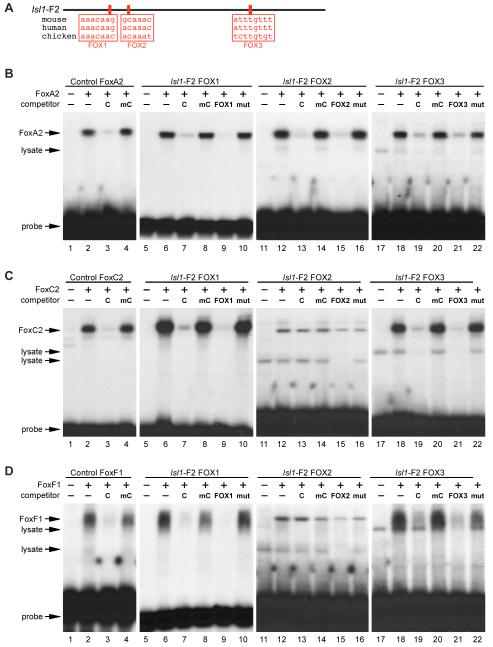
Fig. 4.
The Isl1-F2 enhancer contains three high affinity Forkhead binding sites. (A) ClustalW and TESS analyses, comparing the sequence of the conserved enhancer region from mouse, human, and chick, identified three conserved candidate binding sites (red boxes) for Forkhead transcription factors, denoted as FOX1, FOX2, and FOX3. (B-D) Recombinant FoxA2, FoxC2, and FoxF1 proteins were transcribed and translated in vitro and used in EMSA with a radiolabeled double-stranded oligonucleotide encompassing the three Isl1 FOX sites as well as control FOX sites. Lanes 1, 5, 11, and 17 in panels B-D contains reticulocyte lysate without any recombinant protein. (B) Recombinant FoxA2 efficiently bound to all three Isl1 FOX sites (lanes 6, 12, and 18). Binding was specifically competed by excess unlabeled FoxA2 control probe and by unlabeled self probe, but not by a mutant versions of the probes (FOX1, lanes 7-10; FOX2, lanes 13-16; FOX3, lanes 19-22). (C) Recombinant FoxC2 bound to the FOX1 and FOX3 sites but not to the FOX2 site (lanes 6, 12, and 18) and binding to the FOX1 and FOX3 sites was specifically competed by excess control or self probe, but not by a mutant probes (FOX1, lanes 7-10; FOX3, lanes 19-22). (D) Similarly, recombinant FoxF1 protein bound to the FOX1 (lanes 6-10) and FOX3 (lanes 18-22) sites but not to the FOX2 site (lanes 12-16). Each protein bound efficiently to its bona fide specific control probe and was competed for binding by an excess control probe, but not by a mutant version of the probe (lanes 2-4 in panels B-D). Free probe and lysate-derived mobility shifts are indicated.