Figure 3.
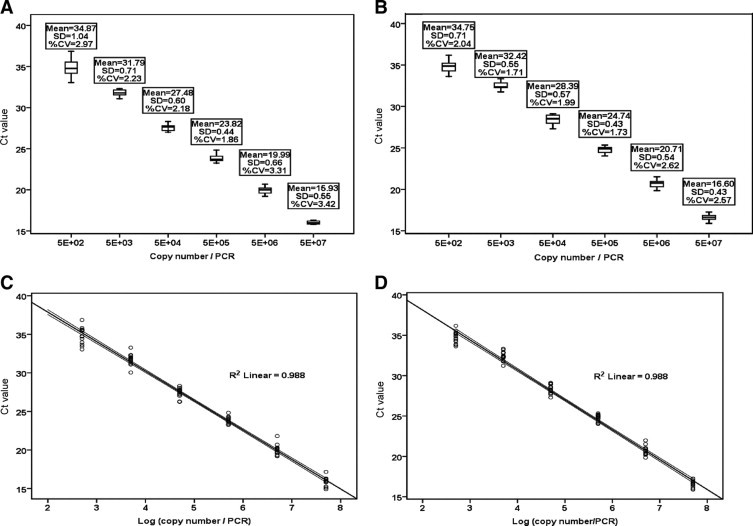
A: Boxplot for Ct values of 15 consecutive runs for the six-point standard curve of the 10-fold serially diluted NP RNA transcript (5 × 102–5 × 107 copies per PCR). Percentage coefficients of variation (%CV) for all six standards were less than 3.5%, and SD, less than 1.5. B: Boxplot for Ct values of 15 consecutive runs for the corresponding six-point standard curve of the 10-fold serially diluted HA RNA transcript (5 × 102–5 × 107 copies per PCR), with %CV for all six standards of less than 3%, and SD, less than 0.8. Boxes in both charts contain the middle 50% of the Ct values, with the upper hinge of the box indicating the 75th percentile of the Ct values, the lower hinge indicating the 25th percentile, and the line in the box denoting the median value of the data. The ‘whiskers’ indicate the minimum and maximum Ct values for each set of 15 replicates. X-Y (scatter) plots of Ct values in 15 consecutive runs against the log10 viral gene copies/PCR were drawn. The best fit lines for the amplification of NP (C) and HA (D) transcripts gave R2 = 0.988 for both.
