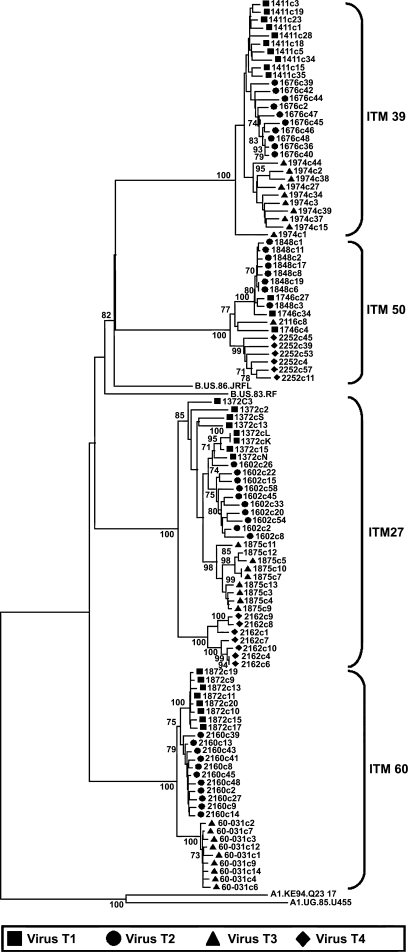
FIG. 5.
Phylogenetic analysis of the sequential HIV-1 sequences obtained from the HIV-1 cohort. Phylogenetic trees were initially constructed with all subtype references available at the Los Alamos Sequence DataBase. All the clones sequenced clustered with subtype B references. Subsequently all the sequences from the patients (ITM27, ITM60, ITM50, and ITM39) were then phylogenetically analyzed with subtype B references and the tree rooted with two subtype A references. The bootstrap values are shown at the internodes. Sequences obtained at sequential visits are shown in sequential order as virus T1, virus T2, virus T3, and virus T4 and represent the visits from which virus was obtained and used in the neutralization studies. Between 1 and 11 clones at each visit were sequenced and analyzed.