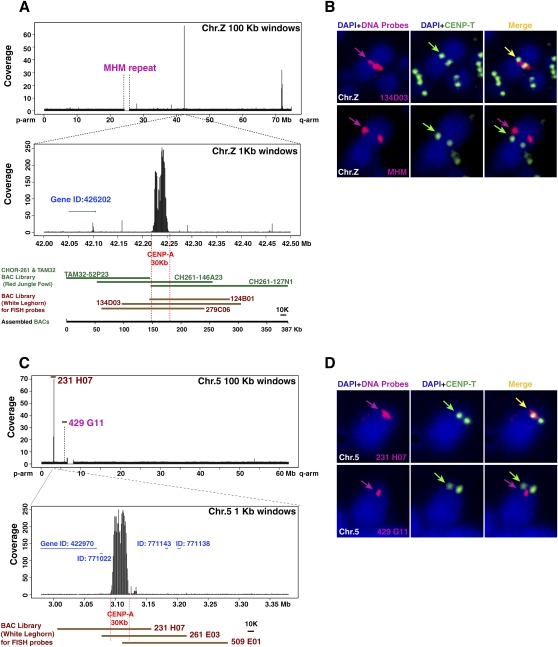
Figure 4.
Centromere region is devoid of tandem-repeated sequences in chromosomes 5 and Z. (A) A distribution of DNA precipitated with CENPA in entire chromosome Z. Major peak is around position of the 42 Mb. High-resolution profile in 1000-bp windows indicates that the binding region of CENPA is restricted in the 30-kb region. Detail sequence information is available in several BAC contigs. As part of sequence information around the 42-Mb position was absent in the NCBI database, we remapped the DNA against BAC contig (387 kb) covering these genomic region and the CENPA associated-DNA was mapped in the 30-kb region, which does not contain tandem-repetitive sequence. Gene ID around CENPA-associated DNA is shown. (B) FISH analysis using a BAC clone (134D03) containing CENPA binding region of chromosome Z (red in top panel). Centromeres are immunostained with anti-CENPT antibodies (green). FISH signals of the 134D03BAC colocalized with centromeres, while FISH signals of the MHM repeat sequence (BamHI 2.2-kb unit) (red in bottom panel) did not localize with CENPT. (C) A distribution of DNA precipitated with CENPA in entire chromosome 5. Major peak is around position of the 3 Mb. High-resolution profile in 1000-bp windows indicates that the binding region of CENPA is restricted in the 30-kb region. Gene IDs around CENPA associated DNA are shown. Clone numbers of BACs are indicated. (D) FISH analysis using a BAC clone (231H07) containing CENPA binding region of chromosome 5 (red in top panel). Centromeres are immunostained with anti-CENPT antibodies (green). FISH signals of the 231H07 BAC colocalized with CENPT, while FISH signals of a BAC clone (429G11) around chromosome 5 satellite repeat sequence (red of bottom panel) did not localize with CENPT.