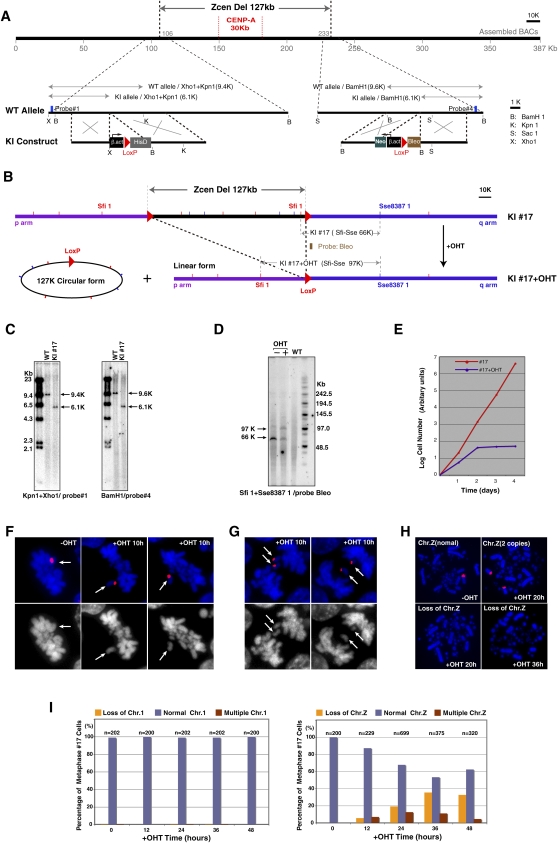
Figure 5.
Deletion of centromere region of chromosome Z results in chromosome instability. (A) To delete entire centromere region of chromosome Z, loxP sequences were inserted into both flanking sites of the centromere region using homologous recombination. CENPA binding region is also shown. (B) Expected genotype after recombination between both loxP sites upon activation of Cre recombinase. The 127-kb region should be removed after addition of OHT. (C) Confirmation of loxP insertions at both flanking sites of the centromere region. Information of restriction map and probes (#1 and #4) for Southern hybridization are shown in A. (D) Confirmation of recombination between both loxP sites. PFGE analysis was performed. Information for the restriction map and the probe (Bleo) for Southern hybridization is shown in B. After recombination a novel 97-kb band is observed. (E) Growth curve of DT40 cells after removal of centromere DNA. (F) Representative images of abnormal metaphase cells in which Z chromosome is not aligned at metaphase plate. Z chromosome was painted with a Z-specific macrosatellite DNA probe (red). (G) Representative images of abnormal anaphase cells in which sister Z chromosomes are left at midzone. Z chromosome was painted with a Z-specific macrosatellite DNA probe (red). (H) Representative images of FISH analysis with a Z-specific macrosatellite DNA probe (red). Cells with two copies of chromosome Z or lacking chromosome Z are observed. (I) Numbers of cells containing chromosome Z after removal of centromere sequence. As a control, numbers of cells containing chromosome 1 were scored. Total cell numbers (n) for the measurements are also shown.