Abstract
The functional complexity of the rice transcriptome is not yet fully elucidated, despite many studies having reported the use of DNA microarrays. Next-generation DNA sequencing technologies provide a powerful approach for mapping and quantifying the transcriptome, termed RNA sequencing (RNA-seq). In this study, we applied RNA-seq to globally sample transcripts of the cultivated rice Oryza sativa indica and japonica subspecies for resolving the whole-genome transcription profiles. We identified 15,708 novel transcriptional active regions (nTARs), of which 51.7% have no homolog to public protein data and >63% are putative single-exon transcripts, which are highly different from protein-coding genes (<20%). We found that ∼48% of rice genes show alternative splicing patterns, a percentage considerably higher than previous estimations. On the basis of the available rice gene models, 83.1% (46,472 genes) of the current rice gene models were validated by RNA-seq, and 6228 genes were identified to be extended at the 5′ and/or 3′ ends by at least 50 bp. Comparative transcriptome analysis demonstrated that 3464 genes exhibited differential expression patterns. The ratio of SNPs with nonsynonymous/synonymous mutations was nearly 1:1.06. In total, we interrogated and compared transcriptomes of the two rice subspecies to reveal the overall transcriptional landscape at maximal resolution.
Rice is one of the most important crops and an excellent monocotyledonous model plant. The global transcriptomes of its different varieties have been surveyed in depth by the genome-wide analysis of full-length cDNAs (FL-cDNAs) and expressed sequence tags (ESTs) (The Rice Full-Length cDNA Consortium 2003; Zhang et al. 2005; Liu et al. 2007). However, the approach of massive-scale cloning and sequencing cDNA or EST libraries is relatively low throughput, high cost, shows inherent cloning bias, and is generally not quantitative. With the availability of rice cDNAs and genome sequences (Goff et al. 2002; Yu et al. 2002; International Rice Genome Sequencing Project 2005), cDNA microarrays and high-density oligonucleotide arrays have been developed to deduce and quantify rice transcriptome profiling on a genome-wide scale (Jiao et al. 2005; Ma et al. 2005; Furutani et al. 2006; Li et al. 2006, 2007; Satoh et al. 2007; Zhang et al. 2008; Wang et al. 2010b). The progress achieved in rice transcriptome analysis has enabled researchers to understand the entire rice gene model sets, expression patterns and their relation to function and regulation, potential transcribed regions, as well as the relationship between global transcription and cytological chromosomes. Despite numerous studies, the inability of microarrays to identify exon splicing junctions restricted us in comprehensively achieving whole-genome transcriptome profiling. It is essential to further globally investigate alternative splicing (AS) patterns, novel transcriptional active regions (nTARs), differentially expressed genes (DEGs), and coding region comparisons between subspecies to improve the annotation of the rice genome.
Recently, the development of the next-generation high-throughput DNA sequencing technologies provided a novel method for both mapping and quantifying transcriptomes (RNA-seq); these new technologies also have the potential to overcome the above-mentioned limitations. RNA-seq technology has been applied to human, yeast, mouse and Arabidopsis, opening the entire transcriptional landscape of gene activity and AS in a high-throughput and quantitative manner (Cloonan et al. 2008; Lister et al. 2008; Mortazavi et al. 2008; Nagalakshmi et al. 2008; Pan et al. 2008; Sultan et al. 2008; Wang et al. 2008; Wilhelm et al. 2008; Filichkin et al. 2010). The updated research focused on integrated comparative analyses of the epigenome and the overall transcriptional output for rice reciprocal hybrids (He et al. 2010). RNA-seq data are highly reproducible, with few systematic discrepancies among technical replicates (Marioni et al. 2008). The latest paired-end tag sequencing strategy of RNA-seq further improves DNA sequencing efficiency and expands short read lengths for better understanding transcriptomes (Fullwood et al. 2009).
Here, we applied the RNA-seq approach to analyze the global transcriptome of rice at the best possible resolution. Compared to previous angles of rice transcriptome research, our study firstly focused on the identification of exon-splicing junctions by relying on the deep sequencing of the transcriptome at single-base resolution by RNA-seq. We developed SPSS (searching positions of splicing sites by RNA-seq data), which we used to detect exon–intron breakpoints from paired-end short reads. We estimate that about half of the rice genes have AS patterns. Second, we identified 15,708 nTARs and validated them randomly by reverse transcription (RT)-PCR. The existing gene annotations were improved as ∼10% of the untranslated region (UTR) boundaries of rice genes have been extended by RNA-seq data. The number of identified SNPs decreased along with the coding sequences and UTRs. Our results also revealed that ∼83% of the gene models could be detected from normal 2-wk seedlings using sequences generated on the Illumina Genome Analyzer II (GAII).
Results
Experimental design
To analyze the functional complexity of the rice transcriptome, we used shotgun sequencing of transcripts to generate randomly distributed reads at base-pair resolution by RNA-seq. We prepared the cDNA according to the protocol of Marioni et al. (2008) with some modifications. Briefly, total RNA was extracted from two-week-old seedlings of Asian cultivated rice Oryza sativa L. ssp indica and japonica varieties, one japonica (Nipp) and two indica (Gla4 and 93-11); the poly(A) mRNA was purified from the RNA and sheared before it was used to generate cDNA. The cDNA was processed and sequenced with the Illumina GAII. We prepared two biological replicates for each variety and sequenced each sample three times (two lanes of 2 × 40 bp, one lane of 2 × 76 bp), split across three runs of the machine.
High-throughput sequencing and mapping of the rice transcriptome
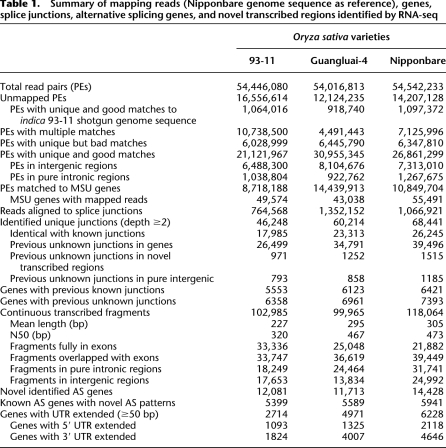
We generated 30.9 million, 27.9 million, and 29.7 million 40-bp-paired-end reads and 23.6 million, 26.1 million, and 24.8 million 76-bp-paired-end reads for the 93-11, Gla4, and Nipp varieties, respectively (Supplemental Table 1). Technical replicates, biological replicates, and 40-bp- vs. 76-bp-paired-end reads of the three varieties were all in agreement referring to gene expression levels where 0.70 < R 2 < 0.88 (Supplemental Fig. 1). We pooled and aligned every individual variety's data against the whole reference genome sequence of japonica cv. Nipponbare (IRGSP v4.0) (Table 1). Tolerances were set to allow at most two mismatches for 40-bp reads and four mismatches for 76 bp in each alignment; reads with multiple alignments were ignored. Moreover, we also extracted 20–36 bp (for 40 bp × 2) and 38–71 bp (for 76 bp × 2) continuous and uniquely mapped reads as candidate exon splicing junction reads. Using these criteria, 38.8%–57.3% of the reads mapped uniquely to a genomic location and 20.2%–24.7% of the reads were filtered as multiple-mapped or low-quality reads (Fig. 1A). Of the unmapped section (22.4%–30.4%), 6.4%–7.7% of the reads could be perfectly matched to the genome shotgun sequence of indica cv. 93-11, indicating where the sequence gaps remain. Based on the rice MSU gene set (version 6.0), 69.3%–73.8% of the mapped reads located to annotated genic regions (plus 1 kb upstream); of these, 3%–4.9% mapped to annotated pure introns. Notably, the remaining 26.2%–30.7% reads mapped to annotated intergenic locations, implying that many putative TARs remain unidentified.
Table 1.
Summary of mapping reads (Nipponbare genome sequence as reference), genes, splice junctions, alternative splicing genes, and novel transcribed regions identified by RNA-seq
Figure 1.
Summary of RNA-seq mapping data. (A) Overall mapping results of paired-end reads (PEs) referring to the Nipponbare genome sequence. (B) Classification of the identified exon–exon junctions based on the known splicing junctions of the MSU annotated gene models. (C) Splicing junctions identified by SPSS and TopHat. (D) RT-PCR validation of 40 randomly selected novel identified exon–exon junctions were carried out by using the total RNAs of Nipponbare. Amplification of the actin fragment in RT-PCR was used as control.
We also aligned each raw data set against the indica cv. 93-11 genome sequence using the same filtering parameters (Supplemental Fig. 2A). More reads (45.0%–62.2%) showed no hits to the reference; 3.1%–5.4% reads were multiple or low-quality hits, and the remaining 30.1%–46.7% reads were filtered as good reads. We further detected the quality value distributions of the mapped and unmapped reads (Supplemental Fig. 2B). The percentage of low-quality value (phred value 2) of the unmapped reads increased to ∼32%, i.e., twofold higher than that of mapped reads. The very low-quality bases interrupt otherwise good and continuous reads, and were therefore filtered by the alignment parameters. It was probably the key reason for the unmapped reads.
Identification of nTARs
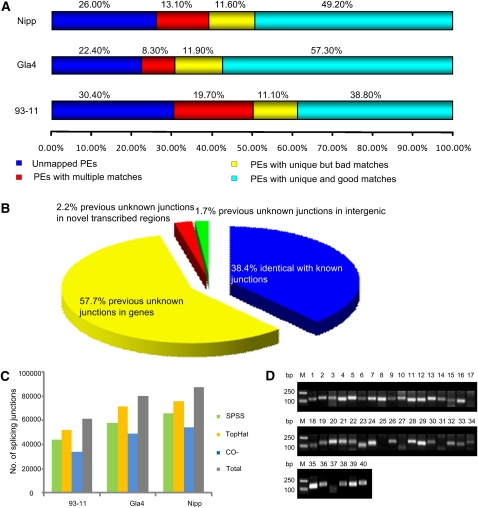
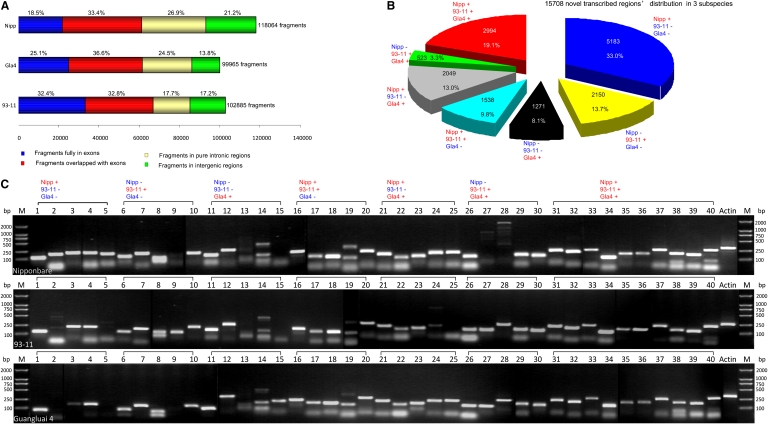
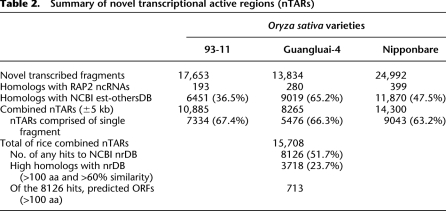
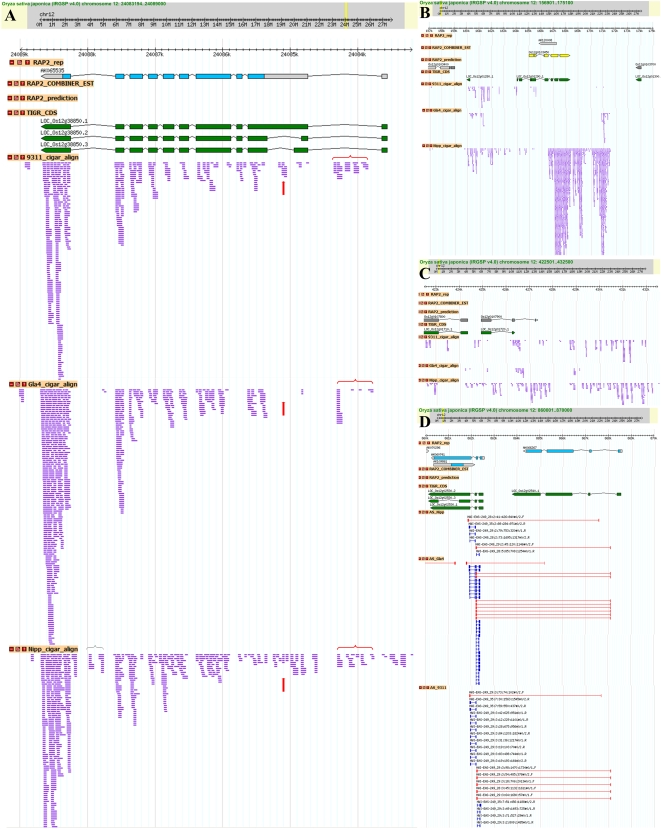
First, we investigated the whole-genome continuous transcribed fragments by mapping the paired-end short reads back to the reference genome. The reads with a continuous mapping length of ≥100 bp and with an average sequencing depth of ≥5 times/bp or a mapping length of 50–100 bp with a depth of ≥10 times/bp were extracted as reliably transcribed fragments. In total, we obtained ∼100,000 transcribed fragments with a mean length of 220–300 bp and ranging in size from 50 to 8610 bp in each variety (Table 1; http://www.ncgr.ac.cn/english/edatabase.htm). Figure 2A showed that 18.5%–32.4% of the fragments fell in annotated exons; 32.8%–33.4% overlapped with exons; 17.7%–26.9% located in pure introns, and the remaining 13.8%–21.2% were completely outside the genic regions (indica 93-11: 17,653; indica Gla4: 13,834; japonica Nipp: 24,992), which were considered putative novel transcribed sequences (Supplemental Table 2). When these novel transcribed sequences were queried against the RAP2 ncRNAs and the updated NCBI est-others database (E-value ≤ 1 × 10−10), ∼2.0% of the fragments showed high similarity to noncoding RNAs, and 36.5%–65.2% had at least one match to known ESTs at >90% sequence identity over the entire length (Table 2). Next, by scanning the genomic location of each novel transcribed fragment, we merged the adjacent fragments (±5 kb) into one transcriptionally active region. In this way, we identified 10,885, 8265, and 14,300 nTARs in 93-11, Gla4, and Nipp, respectively (Table 2). The results showed that >63% of nTARs were surprisingly comprised of single transcribed fragments. This finding is very different from MSU data, where only <20% of the genes are single-exon transcripts. We combined all the novel transcribed fragments and thus identified 15,708 unique rice nTARs. Of the 15,708 unique nTARs, 8126 (51.7%) found no hits in public protein databases when searching against NCBI nrDB (E-value ≤ 1 × 10−6). Only 713 nTARs of the 8126 were predicted open reading frames with >100 amino acids (aa). Of the 7582 matched nTARs, 3718 nTARs had higher similarity to the nrDB entries with >100 aa matches and with >60% similarity. Most of them (3355) were matched to hypothetical proteins, unknown proteins, and expressed proteins; the others were homologous to retrotransposons (57 nTARs), transposons (10 nTARs), polyproteins (10 nTARs), and zinc finger proteins (eight nTARs). We further classified the 15,708 nTARs into seven categories according to the existence of transcribed fragments in the three varieties (Fig. 2B). About 45.2% (7104) of the nTARs were supported by expression data in at least two varieties; of the 7104 nTARs, 2994 were identified in all varieties. Another 2150, 1271, and 5183 nTARs were only identified in 93-11, Gla4, and Nipp, respectively. One example of nTARs is presented in Figure 3C. RT-PCR was carried out to further validate these expression distributions by selecting 40 random nTARs from the seven categories (Fig. 2C).
Figure 2.
Characterization and classification of nTARs. (A) Distribution of continuous transcribed fragments according to the MSU annotated gene models. (B) The 15,708 nTARs were classified in seven categories based on the positive (+) or negative (−) detection of the transcribed fragments in three rice varieties, 93-11, Guangluai 4, and Nipponbare. (C) RT-PCR validation of 40 randomly selected transcripts from the seven categories (indicated at top) of nTARs was carried out using the total RNAs of the three rice varieties, Nipponbare, 93-11, and Guangluai 4, as indicated. Amplification of the actin fragment in RT-PCR was used as control.
Table 2.
Summary of novel transcriptional active regions (nTARs)
Figure 3.
Examples of identified novel AS patterns and nTARs. (A) Novel exons (brackets) and alternative exons (vertical arrows) in the LOC_Os12g38850 transcript among 93-11, Guangluai 4, and Nipponbare are shown. Exons (filled boxes and horizontal arrows) and introns (lines) predicted by gene models (RAP2 and TIGR) were indicated. The RNA-seq short reads were indicated by purple lines. (B) Strong transcriptional activity was detected by the RNA-seq reads (purple lines) in the Nipponbare nearby gene LOC_Os12g01290. (C) nTARs identified by RNA-seq reads (purple lines) in Nipponbare and 93-11 as compared with the gene prediction models. (D) Novel exon–exon splicing junctions were identified in the three varieties.
Extension of gene boundaries
To more precisely define the 5′ and 3′ gene boundaries, we investigated the upstream and downstream regions of all transcripts using the continuous transcribed fragments. In total, 1093, 1325, and 2118 genes were extended at the 5′ end in 93-11, Gla4, and Nipp, by at least 50 bp, whereas 1824, 4007, and 4646 genes were extended at their 3′ end. Only 203, 361, and 536 genes were extended at both ends in 93-11, Gla4, and Nipp (Supplemental Table 3). We further compared the UTR-length distributions for an analysis of Gene Ontology (GO) categories (Supplemental Fig. 3). Transcripts encoding binding proteins had longer 5′ UTRs, whereas the structural molecular activity and catalytic activity genes had shorter 5′ UTRs. The mean UTR lengths of variety 93-11 were shorter than those of Nipp and Gla4 (Supplemental Fig. 3).
Exon splicing junction identification and alternative splicing assessment
In addition to identifying nTARs and gene boundaries, we used the sequencing data to identify novel exons and splicing junctions, and to study AS. We developed SPSS, an approach to scan breakpoints of exon–exon junctions from paired-end short reads. We assessed the efficiency and reliability of this approach by detecting known splicing events. We validated 82.5% of the annotated splicing junctions with 93.0% accuracy. We also discovered the splice junctions using the TopHat software (Trapnell et al. 2009). About 78%–85% of the junctions identified by SPSS were consistent with 66%–71% of the junctions identified by TopHat (Fig. 1C). Collectively, using RNA-seq data, we identified 46,248, 60,214, and 68,441 unique GT–AG splicing junctions (depth of each junction ≥ 2) in 93-11, Gla4, and Nipp with 764,568, 1,352,152, and 1,066,921 paired-end reads, respectively (http://www.ncgr.ac.cn/english/edatabase.htm). Only ∼39% of the exon–exon junctions belonged to annotated splicing sites, whereas >57% were mapped to previously unknown splice variants located in annotated genes (Fig. 1B). Other <5% were identified as novel junctions of unknown transcripts. We randomly selected 40 novel exon–exon junctions for RT-PCR validation (Fig. 1D); of these, 35 gave positive results. An example of novel splicing junctions is presented in Figure 3D. We used the continuous transcribed fragments to find putative novel exons of annotated genes. As described above (Table 1), 18,249 (93-11), 24,464 (Gla4), and 31,741 (Nipp) fragments fell in regions annotated as purely intronic, and 33,747, 36,619, and 39,449 fragments overlapped with known exons. Hence, these fragments can be regarded as fragments of putative novel exons of annotated genes. Combining novel splice junction variants, novel exon fragments, and known AS genes, we identified a total of 17,480, 17,302, and 20,369 AS genes of 93-11, Gla4, and Nipp, respectively (Supplemental Table 4). Of these, ∼70% of the genes were previously unannotated AS genes (Table 1). In other words, at least 41.6%–47.8% of the rice genes were alternatively spliced. An example of a putative alternative splice variant is depicted in Figure 3A. One putative novel exon was identified with solid transcriptional supports between the first and second annotated exons of the gene LOC_Os12g38850 in the three rice varieties.
Differentially expressed genes between two subspecies
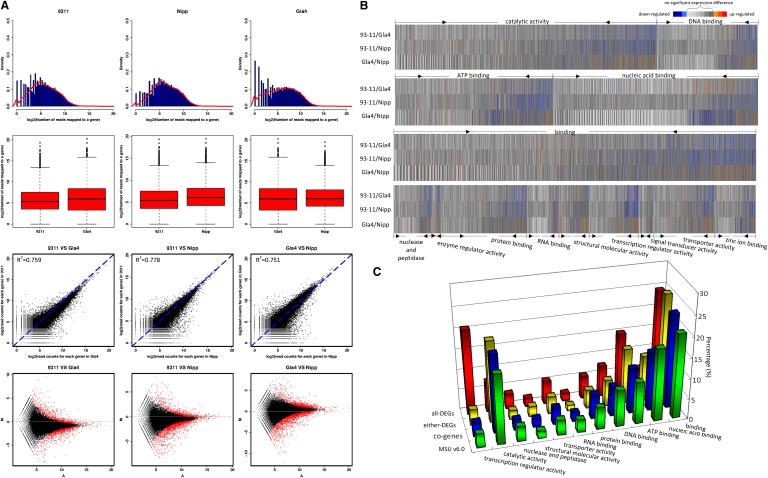
RNA-seq data analyses revealed extensive expression of the whole rice genome (Fig. 4). We further investigated the overall expression by alignment against the MSU gene models and found that 64.7%–83.4% of the annotated genes (49,574 in 93-11, 43,038 in Gla4, and 55,491 in Nipp) were detected by at least one sequence read (Table 1). Overall, there were clear linear relationships in the gene expression levels (0.76 < R 2 < 0.78) among the three varieties (Fig. 5A). The number of reads mapped to different genes was broad and ranged from one to over 300,000 with a median of 23 for 93-11, 42 for Gla4, and 28 for Nipp. Of the mapped genes, 34,738 genes had at least two mapped reads in all the three varieties, assigned as co-genes (Supplemental Table 5). The GO analysis revealed that the genes encoding the binding proteins were enriched in these coexpressed genes (Fig. 5B). The second class of most enriched proteins was related to catalytic activity.
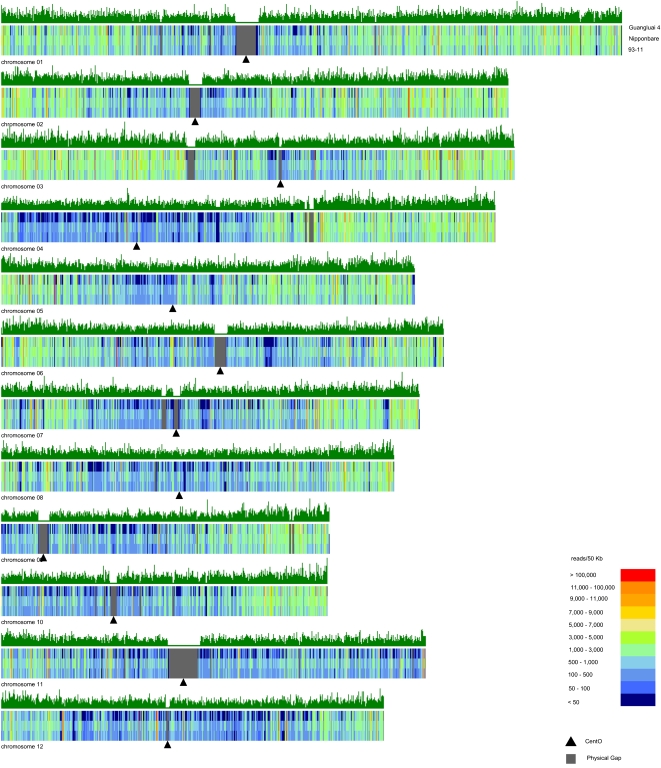
Figure 4.
The genome distribution of transcribed regions in rice. Plots showing the number of mapped paired-end reads that was calculated in 50-kb windows along the 12 rice chromosomes are shown as color-coded vertical bars (see color index).
Figure 5.
Analyses of DEGs between indica and japonica. (A) The top row of graphs show log2 ratios of the expressed genes in 93-11, Nipponbare, and Guangluai 4, respectively. The second row of box-and-whisker plots show log2 transformed values of the expressed genes in the three rice varieties. The third row of graphs show the scatter plot comparing the gene expression levels between 93-11 and Gla4, between 93-11 and Nipp, and between Gla4 and Nipp, respectively. The bottom row of graphs show the genes identified as differentially expressed by Fisher's exact test (red dots). (B) Cluster display of 34,738 coexpressed genes by GO analysis. Differentially expressed genes are shown in red (up-regulated) and blue (down-regulated). Genes with similar expression levels are shown in gray. (C) Genes are categorized on the basis of expression support. (MSU) MSU annotated ; (co-genes) coexpressed genes; (either-DEG) differentially expressed genes in either two varieties; (all-DEG) differentially expressed genes in all three varieties. The percentage of each molecular function for the gene categories is shown.
We measured gene expression levels for further identification of DEGs using the newly developed method (see Methods). On account of sequencing errors and varying sequencing lengths (40 bp vs. 76 bp), the squared correlation coefficient (R 2) across the lanes for samples of each variety was lower than 0.88 (Fig. 5A). Therefore, we computed the appropriate Q-value to eliminate a lane effect of <0.05. At a false discovery rate (FDR) of 1 × 10−22 (for 93-11 vs. Nipp) or 1 × 10−30 (for 93-11 or Nipp vs. Gla4) and an estimated absolute log2-fold change of >1, we identified 3464 genes as reliable DEGs in at least two varieties (assigned as either-DEGs). Functional analysis indicated that they were involved in various protein categories (Fig. 5B). Next, the DEGs of 93-11 vs. Gla4, 93-11 vs. Nipp, and Gla4 vs. Nipp were 1353, 1802, and 2000, respectively. Compared to 93-11 and Nipp, the DEGs of Guangluai 4 showed higher expression of activity in various functions but in nucleic acid binding (Fig. 5B). Eighty genes were expressed differentially in all of rice varieties (assigned as all-DEGs). Of the 80 all-DEGs, we observed that those with transcriptional regulator activity were notably enriched, and those with catalytic activity were reduced (Fig. 5C).
We also measured gene expression levels in reads per kilobase of exon model per million mapped reads (RPKM) (Mortazavi et al. 2008). Referred to this analysis, the DEGs of 93-11 vs. Gla4, 93-11 vs. Nipp, and Gla4 vs. Nipp were identified as 803, 838, and 1028, respectively (Supplemental Table 7). We found that 69.2%–74.0% of these RPKM-based DEGs overlapped with those DEGs that were identified by our method used in this study.
Identification of SNPs and comparative analysis
RNA-seq technology also holds the promise of quickly and reliably detecting single-nucleotide polymorphisms (SNPs). We made transcriptome comparisons between two rice subspecies, indica and japonica, using SNPs detected from 93-11 and Gla4 RNA-seq data. When referring to the japonica Nipponbare genome sequences, we identified 67,011 SNPs between 93-11 and Nipp, and 64,481 SNPs between Gla4 and Nipp. We also identified 26,480 SNPs between the Nipp transcripts and the Nipp genome sequence. In addition, 8729, 2825, and 14,263 relatively reliable heterozygous SNPs were identified in 93-11, Gla4, and Nipp, respectively (http://www.ncgr.ac.cn/english/edatabase.htm). Only half of these SNPs (32,920 SNPs from 93-11 vs. Nipp and 39,809 SNPs from Gla4 vs. Nipp) were located in 16,597 annotated gene models. High SNP frequencies appeared at the first 200 bp upstream of the translation start site, at the initial 800 bp of coding sequences, and 400 bp downstream from the stop codon, respectively (Supplemental Fig. 4A). About 60.8% of the SNPs were located in coding regions, 29.1% were found in the 3′ UTR and 10.1% in the 5′ UTR. For 93-11 vs. Nipp, the total numbers of nonsynonymous SNPs and synonymous SNPs were 8622 and 9144, respectively. For Gla4 vs. Nipp, the total numbers of nonsynonymous SNPs and synonymous SNPs were 9684 and 10,303, respectively. Therefore, the ratio of nonsynonymous vs. synonymous SNPs was nearly 1:1.06. Most of the nonsynonymous SNPs belonged to the first and second nucleotides of the codon unit, while the majority of synonymous SNPs were the third base. We found that proteins with transcription regulator activity, zinc ion binding, and chromatin binding had more variations between indica and japonica as their encoding genes had more nonsynonymous SNPs; proteins with structural molecular activity, transporter activity, signal transducer activity, nutrient reservoir activity, and polymerase activity were more conserved as their encoding genes had more synonymous SNPs (Supplemental Fig. 4B).
Discussion
Our results demonstrated the global transcriptome profiling of two rice subspecies, indica and japonica, using ultra-high-throughput sequencing technology. We evaluated the applicability of the method to various analyses, including novel transcribed region discovery, splicing site detection, gene expression assessment, and SNP identification.
The data set of 42 million (40 bp × 2) and 35 million (76 bp × 2) uniquely mapped reads provided rich resources for improving gene annotations across the rice genome. Our results showed that a quarter of these reads mapped in intergenic regions, which generated 15,708 nTARs. Randomly RT-PCR validated their expression among RNA samples and hence provides a reliable estimation of additional transcribed genomic loci beyond the annotated genes. Half of these nTARs could be potential noncoding RNAs for the following reasons: (1) Over 63% nTARs belonged to single transcribed fragments with a mean length of ∼220 bp within its ±5-kb genome sequence ranges (Referring to MSU annotated data, 99.76% of intron sizes were <5 kb.); (2) 47.2% of RAP2 noncoding RNAs could be mapped to ∼2.0% of the novel transcribed fragments; (3) 51.7% of nTARs gave no hits to the NCBI nrDB database.
In addition, the massively produced reads made it possible to identify unique transcripts of each individual variety. Our results indicated that 2150, 1271, and 5183 nTARs were uniquely identified in 93-11, Gla4, and Nipp, respectively. Some of the unique nTARs of each variety could be validated by RT-PCR since one-fifth to two-fifths of the primer products gave a positive signal only in one variety as expected (“Nipp+, 93-11−, Gla4−”; “Nipp−, 93-11+, Gla4−”, and “Nipp−, 93-11−, Gla4+” in Fig. 2C). The RT-PCR results suggested that the expression of many of these genes was not detected under the experimental conditions used in this investigation.
Our data also provided evidence for thousands of extended or newly identified 5′ and 3′ UTRs. The extended 5′ UTRs imply more positions for promoters and alternative promoters, which might be involved in the interactions with various regulatory factors. More alternative promoters of transcripts encoding ATP binding, protein binding, and nucleic acid binding existed because their genes had longer 5′ UTRs. The extended 5′ UTRs and 3′ UTRs would be valuable for determining the intact gene boundaries, which is also helpful for finding genomic loci with transcripts on both strands (termed natural antisense transcripts [NATs]). In addition, the extended 3′ UTRs might be relevant to their possible roles in microRNA-mediated control of translation and post-transcriptional RNA regulation. Moreover, to build a complete and precise map of UTR-extended sequences (especially for distinguishing sense-antisense gene boundaries), strand-specific RNA-seq (ssRNA-seq) data are required.
AS is an efficient way for genomes to code for more transcripts. The identification of AS patterns of rice genes will be helpful to better understand the mechanism of their transcription control. Previous studies estimated that 20%–30% of the rice genes with EST/cDNA data undergo AS (Campbell et al. 2006; Wang and Brendel 2006). Here, our RNA-seq data, combined with the MSU annotated AS patterns, indicate that at least 48% of the genes in rice japonica Nipponbare are alternatively spliced. The updated RNA-seq data of Arabidopsis indicate that ∼42% of the intron-containing genes are subject to AS (Filichkin et al. 2010). It revealed that AS in plants is considerably more widespread than previously predicted. As expected, the consensus GT–AG intron boundaries constitute the highest proportion (>99.0%). Deeper ultra-high-throughput sequencing, longer paired-end reads, lower sequencing errors of rice transcriptome data, and more RNA samples from different tissues at various developmental stages are essential for further comprehensively analyzing rice AS patterns to produce a more integrated splicing pattern map. Also, ssRNA-seq technology will be additionally informative. In Figure 3B for example, without ssRNA-seq data, it would be difficult to determine whether the transcript identified in Nipp is an improved annotation of gene LOC_Os12g01290, an AS pattern, or an individual novel transcript in the opposite orientation. Moreover, experimental confirmation would be vital for further validating these computational predictions and understanding their biological functions.
Finding all the exon–exon splicing junctions is a challenging task as the RNA-seq reads are short; sequencing errors always occur, and many splice junctions may be spanned by very few reads. Here, our SPSS program showed 82.5% efficiency with 93.0% accuracy. The mis- and error-identified sites are probably due to the following reasons: (1) Part of the reads could not be mapped contiguously to the reference because of sequencing errors; (2) sequences of some splicing junctions were ignored when they gave multiple hits to the reference; (3) with the existence of simple sequence repeats (SSRs), the alignment results of reads might not be the genuine locations. The efficiency of junction discovery was not as high as that of TopHat. However, SPSS could discover some additional splice junctions. The discrepancies were observed probably because different alignment software packages were used and different parameters were applied by SPSS and TopHat.
High-throughput sequencing is a powerful and quantitative method for the in-depth analysis of transcriptomes at a high resolution. Consistent with previous reports (Li et al. 2006), the RNA-seq data provided a maximal resolution map of the different transcriptional activities associated with heterochromatin and euchromatin in the rice genome (Fig. 4). The different expression levels of the transcripts could be due to slight differences in the rate of plant development; other biological differences probably also existed between the varieties. Our analyses of DEGs in 93-11, Gla4, and Nipp provided an alternative aspect to plant genomic comparative research. Using the χ2 goodness-of-fit test, the RNA-seq results could be reliably measured to identify DEGs in an unbiased manner by eliminating the lane effect. Less rounds of amplification for input RNA with high-throughput sequencing would presumably give a more precise estimation of the gene expression level.
We found 75,740, 67,306, and 40,743 SNPs (including reliable heterozygous SNPs) between the transcripts of 93-11 and Nipp, between the transcripts of Gla4 and Nipp, and between the transcripts of Nipp and Nipp, respectively. This represents a greater number of SNPs than expected. Possible reasons for these SNPs are as follows: They may reflect the real SNPs between rice subspecies; they may be affected by RNA editing; they likely belonged to sequence errors in spite of sequence quality filtration; they may be derived from the mapping error, or they were simply mistaken by a reference sequencing error. It should be pointed out that, according to present data, we could not clearly determine how many SNPs are doubtless due to RNA editing. Using both genomic DNA and RNA from the same organism of the same variety for deep-sequencing with higher base quality might be a better way to detect and analyze RNA editing.
To date, we only extracted uniquely mapped reads (according to reference IRGSP v4.0) for this study. Another 8.3%–19.7% of the multiple mapped reads in our RNA-seq data remained unanalyzed. Most of the multi-reads could be attributed to known duplicated genes and segmental duplications. In addition, 22.4%–30.4% of the reads could not be mapped to the Nipponbare genome sequence. The main reasons for these unmapped reads are sequencing errors (Supplemental Fig. 2B) and the uneven quality of the sample preparations; another reason is the physical gap of the reference—6.4%–7.7% reads of this section could be matched to the 93-11 genome sequence; further reasons include reference errors and the defined mapping criterions. When we used the indica 93-11 genome sequence as a mapping reference, the unmapped section was ∼45%–62%, indicating that the 93-11 genome sequence is incomplete.
In summary, the RNA-seq approach was proven as an efficient method for transcriptome profiling analyses. It can be widely applied for various research purposes, ranging from identifying novel transcribed active regions, splicing sites, and AS patterns for measuring the mRNA expression for detecting DEGs of different samples when comparing subspecies.
Methods
Plant materials and growth conditions
Seeds from the cultivated rice subspecies Oryza sativa ssp. japonica Nipponbare, indica 93-11, and Guangluai 4 were grown in soil under 12-h-light/12-h-dark, at 28°C in a greenhouse. Shoots from 2-wk-old seedlings were harvested for RNA isolation.
cDNA preparation and library construction for Illumina sequencing
We prepared the cDNA according to a protocol (Marioni et al. 2008) with some modifications. Genomic DNA was digested using DNase (New England Biolabs), and total RNA was isolated using the TRIzol reagent (Invitrogen). The OligoTex mRNA mini kit (Qiagen) was used to purify poly(A) mRNA from the total RNA samples. Then the mRNA was fragmented by the RNA fragmentation kit (Ambion); the first cDNA strand was synthesized using random hexamer primers, and the second strand cDNA was synthesized next.
For high-throughput sequencing, the sequencing library was constructed by following the manufacturer's instructions (Illumina). Fragments of ∼300 bp were excised and enriched by PCR for 18 cycles. The products were loaded onto flow cell channels at a concentration of 2 pM for paired-end 40 bp × 2 or 76 bp × 2 sequencing. The Illumina GA processing pipeline v0.2.2.6 was applied for image analysis and base calling.
Data analysis
Sequence alignments were performed by SSAHA2 v2.3 (Sequence Search and Alignment by Hashing Algorithm) from the software package pileup_v0.5 (http://www.sanger.ac.uk/Software/analysis/SSAHA2/). The “sol2sange” pipeline in MAQ (Li et al. 2008) was used to convert the Illumina FASTQ to the Sanger standard FASTQ format to match the SSAHA2 input file format. For calculating the sequence depth of each nucleotide on pseudomolecules and further determining the ranges of the continuous transcribed fragments, the “ssaha_pileup” pipeline in pileup_v0.5 was executed with the parameter “-solexa 1 -cons 1 -trans 1” (ftp://ftp.sanger.ac.uk/pub/zn1/ssaha_pileup/). Perl scripts were written for various analysis purposes. Similarity searches of putative novel transcripts were performed using BLASTX v2.2.14 (Altschul et al. 1997) with an E-value < 1 × 10−6. The functional classification of the genes referred to the GO terms attached in InterPro database (Apweiler et al. 2001).
Database application
The following databases were used for data analysis in our research: IRGSP pseudomolecules of rice japonica cv. Nipponbare (Build 4.0 and 5.0, http://rgp.dna.affrc.go.jp/IRGSP/Build4/build4.html), BGI rice indica cv. 93-11 contigs, annotated gene models and chromosome sequences (http://rice.genomics.org.cn/rice/link/download.jsp), NCGR rice indica cv. Guangluai 4 ∼22.1 Mb genome sequence (http://www.ncgr.ac.cn/chinese/databasei.htm), rice MSU data including genes, cDNAs, 3′ UTRs, 5′ UTRs, and introns and exons (release 6.0, ftp:ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.0). Here, we used a total of 66,540 rice MSU cDNA sequences that could be accurately mapped to IRGSP v4.0. The RAP2 noncoding RNA sequence data (http://rapdb.dna.affrc.go.jp/), NCBI GenBank est-others and nrDB (ftp://ftp.ncbi.nih.gov), as well as the InterPro database (ftp://ftp.ebi.ac.uk/pub/databases/interpro/) were also downloaded for data analyses.
Mapping reads to genome data and annotated transcripts
The parameters of SSAHA2 were set as “-pair 50, 6000,” “-outfile abnormal.out,” and “-mthresh 15.” From the primary searching results, reads that aligned to multiple genomic locations were ignored. Of the uniquely mapped reads, tolerances were set to allow at most two mismatches for 40-bp reads and four mismatches for 76 bp for either alignment. At the same time, 20–36-bp continuous mapped reads for the 40-bp paired-end reads, and 38–71-bp continuous mapped reads for 76-bp paired-end reads were also extracted as candidate exon–exon junction reads.
Paired-end reads that were mapped uniquely to the reference were further aligned to exons retrieved from MSU v6.0. The number of reads that were fully located in exons was counted. The expression level for each gene was determined as the sum of all hits in exons.
Identification of novel transcribed active regions
The pure intergenic regions (excluding all UTRs, exons, and introns) were bordered using the rice MSU cDNA data and the Nipponbare pseudomolecules. Stretches of contiguous expression in intergenic regions were further identified. Those with a continuous mapping length of ≥100 bp with an average sequencing depth of ≥5 times/bp or a mapping length of 50–100 bp with a depth of ≥10 times/bp were required for putative novel transcribed fragments. When allowing for 95.4% of the rice gene sizes to be <8 kb and 99.76% intron sizes to be <5 kb (referred to rice MSU data), we combined novel transcribed fragments which located in the adjacent genomic regions (±5 kb) into one individual transcribed unit. Novel transcribed units were assigned as nTARs and were searched against nrDB and were randomly selected for further validation by RT-PCR.
Gene boundary determination using continuous transcribed fragments
The continuous transcribed regions were merged together if the gap length between two transcriptional fragments was <30 bp (as only ∼0.1% of rice genes had an intron length of <30bp). Then, the rice MSU gene boundaries were compared to the combined transcribed regions to determine the potential extended gene boundaries by screening for a break in the transcribed region overlapping the annotated UTRs. The UTR was considered extended if the upstream/downstream sequence length of its overlapping transcribed fragments was >50 bp.
Detection of splicing sites
The initial SSAHA2 results generated a set of sequence reads that partially matched the reference genome sequence and used potential spliced reads. We selected candidate trans-reads for further validation using the following criteria: For 40-bp paired-end reads, we extracted those with 20–36 bp continuously and uniquely matched to the reference; for 76-bp paired-end reads, we extracted those with 38–71 bp. We developed SPSS, written in C++, to detect the breakpoints of splicing sites. The rationale of this program is briefly described here. First, the location of the putative trans-reads on the genome sequence needed to be determined; it was also possible to recognize the orientation of this read. Second, we searched the unmapped part of the trans-read against that of 50 bp to 6 kb limit for the maximum sequence range from the upstream or downstream sequence, according to its location and orientation on the reference sequence (if necessary, the sequence should be reversed). Third, we filtered those matches by scanning the GT/AG (reverse, CT/AC) introns of each matched trans-read. Finally, a depth of each identified junction ≥2 was required. Another GC/AG intron rule was also tested to detect AS patterns. We also applied TopHat, an available software package, to discover splice junctions to evaluate the efficiency of SPSS (Trapnell et al. 2009).
To validate the novel splicing junctions, we designed 40 primers and used the total RNAs of Nipponbare for RT-PCR (Supplemental Table 6).
Differentially expressed gene assessment
We detected gene expression levels for further DEG identification using the following calculation: (Matched PE reads per gene/total matched PE reads of one variety) × (total matched PEs in all three varieties/3). We then identified DEGs from different varieties (93-11, Gla4, and Nipp) according to an R package named “DEGseq” (Wang et al. 2010a). The Pearson's χ2 test was applied to assess the lane effect. For each gene, the P-value and Q-value were computed. Then, the significance threshold to control the FDR at a given value was calculated. The fold changes were also estimated within the R statistical package. We also applied reads per KB per million reads (RPKM) to detect gene expression levels (Mortazavi et al. 2008).
SNP calling
For SNP detection, the “ssaha_pileup” pipeline in pileup_v0.5 was executed for the high-quality reads of 93-11 and Gla4 (described in section Mapping Reads to Genome Data and Annotated Transcripts) with the parameter “-solexa 1”. SNPs with low-quality sequences (SNP_score in output file < 20) and low sequence depth (N_reads in output file < 5) were discarded. Then, homozygous SNPs and reliable heterozygous SNPs were identified according to their SNP quality score. Furthermore, only the isolated SNPs (i.e., no adjacent and sequential SNPs) were identified as reliable SNPs between indica and japonica.
Novel transcribed active region validation by RT-PCR.
Forty primers were designed for validating the nTARs (Supplemental Table 6). RNA was extracted using the TRIzol reagent (Invitrogen) and RT-PCR was performed according to the manufacturer's instructions.
Acknowledgments
We thank Tong Zhu for valuable discussions. This work was supported by the Ministry of Science and Technology of China (grant no. 2006AA10A102), the Ministry of Agriculture of China (grant nos. 2008ZX08009-002 and 2008ZX08012-002), the Chinese Academy of Sciences (grant no. KSCX2-YW-N-024), and the National Natural Science Foundation of China (grant no. 30821004).
Footnotes
[Supplemental material is available online at http://www.genome.org. The RNA-seq data from this study have been deposited in the EMBL Sequence Read Archive (SRA) under accession no. ERA000212 (http://www.ebi.ac.uk/ena/data/view/ERA000212) and are available in a genome browser at http://www.ncgr.ac.cn/rrs. The sequence data set of continuous transcribed fragments, the detailed list of identified splicing junctions, all identified SNP lists, the SPSS binary code, and Perl scripts are freely available at http://www.ncgr.ac.cn/english/edatabase.htm.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.106120.110.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res 29: 37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR 2006. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 7: 327 doi: 10.1186/1471-2164-7-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC 2010. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Wei CL, Liu ET, Ruan Y 2009. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res 19: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Sukegawa S, Kyozuka J 2006. Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J 46: 503–511 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F, et al. 2010. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project 2005. The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Jia P, Wang X, Su N, Yu S, Zhang D, Ma L, Feng Q, Jin Z, Li L, et al. 2005. A tiling microarray expression analysis of rice chromosome 4 suggests a chromosome-level regulation of transcription. Plant Cell 17: 1641–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang X, Stolc V, Li X, Zhang D, Su N, Tongprasit W, Li S, Cheng Z, Wang J, et al. 2006. Genome-wide transcription analyses in rice using tiling microarrays. Nat Genet 38: 124–129 [DOI] [PubMed] [Google Scholar]
- Li M, Xu W, Yang W, Kong Z, Xue Y 2007. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol 144: 1797–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lu T, Yu S, Li Y, Huang Y, Huang T, Zhang L, Zhu J, Zhao Q, Mu J, et al. 2007. A collection of 10,096 indica rice full-length cDNAs reveals highly expressed sequence divergence between Oryza sativa indica and japonica subspecies. Plant Mol Biol 65: 403–415 [DOI] [PubMed] [Google Scholar]
- Ma L, Chen C, Liu X, Jiao Y, Su N, Li L, Wang X, Cao M, Sun N, Zhang X, et al. 2005. A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res 15: 1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni J, Mason C, Mane S, Stephens M, Gilad Y 2008. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- The Rice Full-Length cDNA Consortium 2003. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Satoh K, Doi K, Nagata T, Kishimoto N, Suzuki K, Otomo Y, Kawai J, Nakamura M, Hirozane-Kishikawa T, Kanagawa S, et al. 2007. Gene organization in rice revealed by full-length cDNA mapping and gene expression analysis through microarray. PLoS ONE 2: e1235 doi: 10.1371/journal.pone.0001235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg S 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Brendel V 2006. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci 103: 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X 2010a. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138 [DOI] [PubMed] [Google Scholar]
- Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J, et al. 2010b. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J 61: 752–766 [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Zhang J, Feng Q, Jin C, Qiu D, Zhang L, Xie K, Yuan D, Han B, Zhang Q, Wang S 2005. Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J 42: 772–780 [DOI] [PubMed] [Google Scholar]
- Zhang HY, He H, Chen LB, Li L, Liang MZ, Wang XF, Liu XG, He GM, Chen RS, Ma LG, et al. 2008. A Genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol Plant 1: 720–731 [DOI] [PubMed] [Google Scholar]