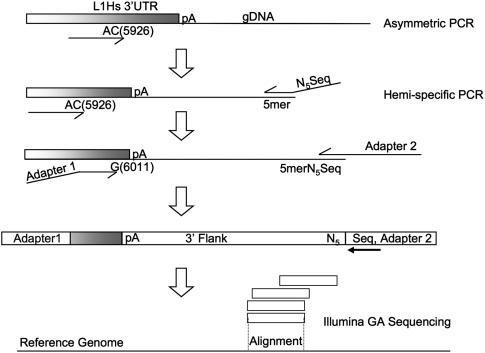
Figure 1.
Hemi-specific PCR scheme to amplify 3′ flanking regions of human-specific LINE-1 insertion sites. The first five cycles of PCR enrich for sequences containing human-specific L1 sequences via primer extension with the single primer pictured above. The AC and G nucleotides in the primers for L1 are diagnostic for the human-specific subfamily for this element. After enrichment for human-specific L1 flanks, a degenerate primer is added that has a specified 5-mer at the 3′ end preceded by five degenerate bases (NNNNN) and a sequencing primer used for the Illumina Genome Analyzer. Eight different reactions are performed, each with a different specified 5mer. The next round of PCR enriches for human-specific L1 3′ flanks with another primer complementary to the L1 and adds the necessary adapter sequences via primer overhangs. The resulting products from each 5-mer are mixed and sequenced on the Illumina Genome Analyzer platform. Following sequencing and initial processing, tags representing the 3′ flanks of human-specific L1 insertions are aligned to the human reference genome (hg18).