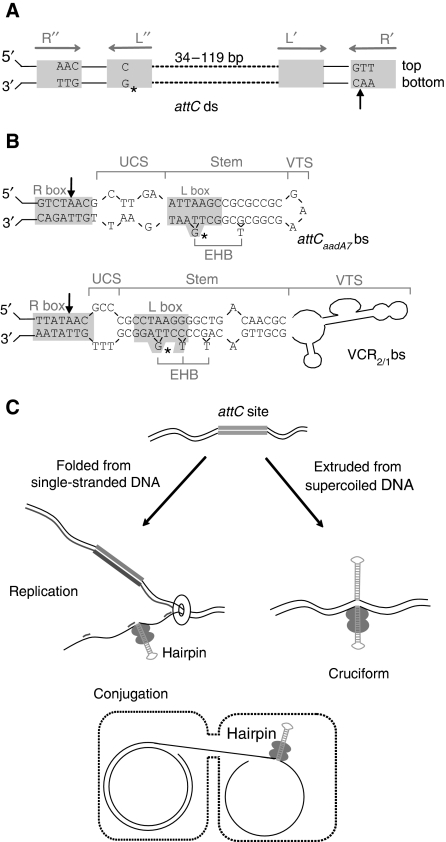
Figure 1.
attC recombination sites and model for the attC folding. (A) Schematic representation of a double-stranded (ds) attC site. Inverted repeats R″, L″, L′ and R′ are indicated by grey boxes. The dotted lines represent the variable central part. The conserved nucleotides are indicated. Asterisks (*) show the conserved G nucleotides, which generate extrahelical bases (EHB) in the folded attC site bottom strand (bs). The black arrow shows the cleavage point. (B) Secondary structures of the attCaadA7 and VCR2/1 sites bottom strands (bs). Structures were determined by the UNAFOLD online interface at the Institut Pasteur. The four structural features of attC sites, namely, the unpaired central segment (UCS), the EHB, the stem and the variable terminal structure (VTS) are indicated. Black arrows show the cleavage points. Primary sequences of the attC sites are shown (except for the VTS of the VCR2/1 site). (C) Cellular processes possibly allowing the attC site folding. The different possible cellular pathways allowing proper folding of the attC site bottom strand are shown: from single-stranded DNA during replication and conjugation, and cruciform extrusion from supercoiled double-stranded DNA. IntI monomers are represented as grey ovals.