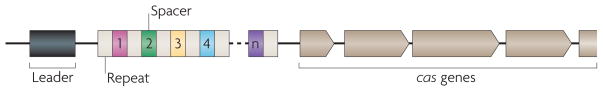
Figure 1. Features of CRISPR loci.
Typically, clustered, regularly interspaced short palindromic repeats (CRISPRs, white boxes) are preceded by a leader sequence (black box) that is AT-rich but otherwise not conserved. The number of repeats can vary substantially, from a minimum of two to a few hundred. Repeat length, however, is restricted to 23 to 50 nucleotides. Repeats are separated by similarly sized, non-repetitive spacers (coloured boxes) that share sequence identity with fragments of plasmids and bacteriophage genomes and specify the targets of CRISPR interference. A set of CRISPR-associated (cas) genes immediately precedes or follows the repeats. These genes are conserved, can be classified into different families and subtypes, and encode the protein machinery responsible for CRISPR activity.