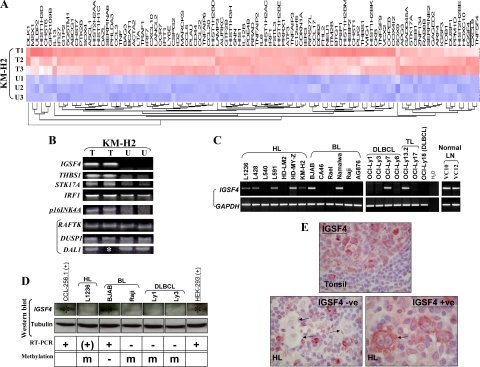
Figure 1.
A: Heat map of genes up-regulated in KM-H2 cells following demethylating treatment with Aza. T1–3: treated samples; U1–3: controls. IGSF4 is underlined. B: Validation of microarray results by RT-PCR of selected genes up-regulated in KM-H2 cells after Aza treatment (T) compared with untreated cells (U). Upregulation of mRNA for IGSF4, THBS1, STK17A and IRF1, and unchanged expression of RAFTK, DUSP1, and DAL1 are shown. p16INK4A was not picked up by the microarray screening, however its expression was up-regulated after Aza treatment. Asterisk indicates that no RNA left in one sample. C: IGSF4 down-regulation in HL and NHL cell lines by RT-PCR. RT-PCR analysis showed that when compared with two normal lymph nodes, IGSF4 expression was completely lost or reduced in 6/7 HL, 4/5 DLBCL, and 4/6 BL cell lines. D: IGSF4 protein expression correlated with its mRNA expression, absent in methylated cell lines: one HL cell line (L1236), one BL (Raji), and two DLBCL cell lines (Ly1 and Ly3). HEK293 and CCL-256.1 were used as positive controls. Asterisk indicates the +v band for IGSF4 protein. E: Immunohistochemistry with an IGSF4-specific monoclonal antibody in normal lymphoid tissues and HL. Upper panel shows normal expression of IGSF4 protein in GC B cells. In contrast, HRS cells in most HL cases lacked expression or showed lower levels of expression. A representative example of the absence of IGSF4 expression is shown in the lower left panel. HRS cells are indicated with arrows. A minority of cases showed normal expression of IGSF4 in HRS cells (lower right panel). These results were confirmed using two separate chicken polyclonal antibodies directed to IGSF4 (data not shown). See Table 4 for correlation with MSP analysis.