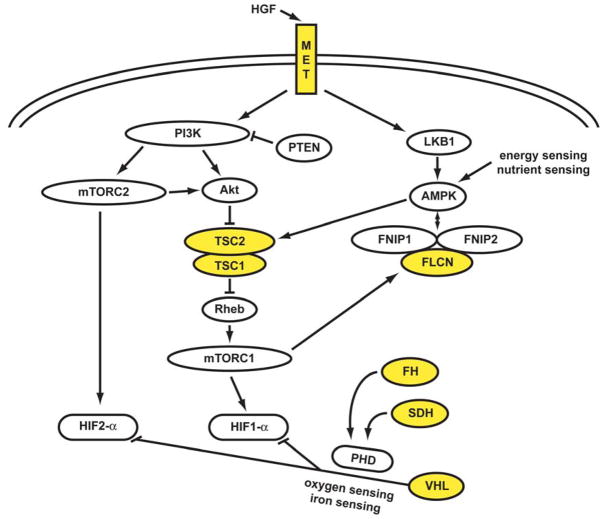
Figure 1. The genetic basis of kidney cancer: a metabolic disease.
The genes known to cause kidney cancer, VHL, MET, FLCN, FH, SDH, TSC1 and TSC2 share the common feature that each is involved in oxygen, iron, energy and/or nutrient sensing pathways. Kidney cancer is fundamentally a metabolic disease. VHL targets HIF-1α and HIF-2α for ubiquitin-mediated degradation through an oxygen and iron sensing mechanism. The FLCN/FNIP1/FNIP2 complex binds AMPK, the primary energy sensor in the cell, and FLCN is phosphorylated by a rapamycin-sensitive kinase (i.e.,mTORC1). TSC1/TSC2 are phosphorylated by the LKB1/AMPK cascade and help mediate the cell’s response to energy/nutrient sensing. Fumarate hydratase and succinate dehydrogenase are TCA cycle enzymes. When fumarate hydratase or succinate dehydrogenase are deficient, the function of the TCA cycle is impaired and the cell is dependent on glycolysis for energy production. Inactivation of fumarate hydratase or succinate dehydrogenase impairs PHD function and represents a VHL-independent mechanism for dysregulation of HIF degradation. Increased HIF levels lead to increased GLUT1 which enables transport of glucose for ATP production.
Abbreviations: folliculin interacting protein 1 (FNIP1) and folliculin interacting protein 2 (FNIP2), HIF prolyl hydroxylase (PHD). Adapted from Linehan, et al.(1)