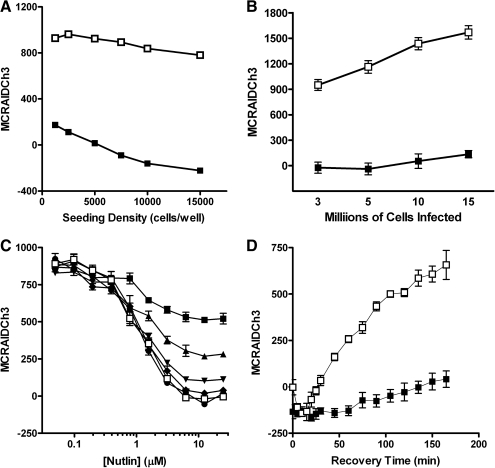
Fig. 3.
Optimization of the p53-hDM2 protein–protein interaction biosensor (PPIB) high-content screening assay. (A) Cell seeding density. p53-hDM2 PPIB adenovirus-infected cells were seeded at the indicated cell densities in the wells of 384-well Greiner collagen-coated assay plates, cultured overnight at 37°C, 5% CO2, and 95% humidity, and were then treated for 90 min with either 0.5% dimethyl sulfoxide (DMSO) (□) or 10 μM Nutlin-3 in 0.5% DMSO (▪) prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. (B) Scalability of the p53-hDM2 PPIB adenovirus coinfection process. U-2 OS cells ranging between 3 × 106 and 1.5 × 107 cells in 1.5 mL of media were coincubated with the manufacturer's recommended volumes of the p53-hDM2 PPIB adenoviruses for 30 min. Coinfected cells were then seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, cultured overnight at 37°C, 5% CO2, and 95% humidity, and were then treated for 90 min with either 0.5% DMSO (□) or 10 μM Nutlin-3 in 0.5% DMSO (▪) prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. (C) Time course of Nutlin-3 disruption. U-2 OS cells were coinfected with the p53-hDM2 PPIB adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Cells were then treated with the indicated concentrations of Nutlin-3 and then fixed and stained with Hoechst at 5 min intervals up to 30 min total exposure time; (▪) 5 min, (▴) 10 min, (▾) 15 min, (♦) 20 min, (●) 25 min, and (□) 30 min. (D) Reversibility and stability of the p53-hDM2 PPIB assay signal window. U-2 OS cells were coinfected with the p53-hDM2 PPIB adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Cells were then treated with 10 μM Nutlin-3 for 90 min. Half the wells were then washed 3 times with fresh McCoy's 5A medium (□) and the remainder were left untouched (▪). Cells were incubated further at 37°C, 5% CO2, and 95% humidity for the indicated times prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. Images from 3 fluorescent channels were sequentially acquired on the ArrayScan VTI platform using a 10× 0.3 NA objective with the XF93 excitation and emission filter set and were analyzed with molecular translocation (MT) image analysis algorithm. The mean circle ring average intensity difference in channel 3 (MCRAID-Ch3) data derived from the MT image analysis algorithm is a measure of the relative distribution of the hDM2-red fluorescent protein within the nucleus and the cytoplasmic regions and was used as the primary indicator of the interactions between p53 with hDM2. The data are presented as the mean MCRAID-Ch3 values ± SD from (A) n = 32 wells, (B) n = 96 wells, (C) n = 4 wells, and (D) n = 6 wells. The lines connecting the data were plotted using Graphpad Prism software 4.03. Data from a single representative experiment of 3 or more experiments are presented in A–C, and from a single experiment in D.