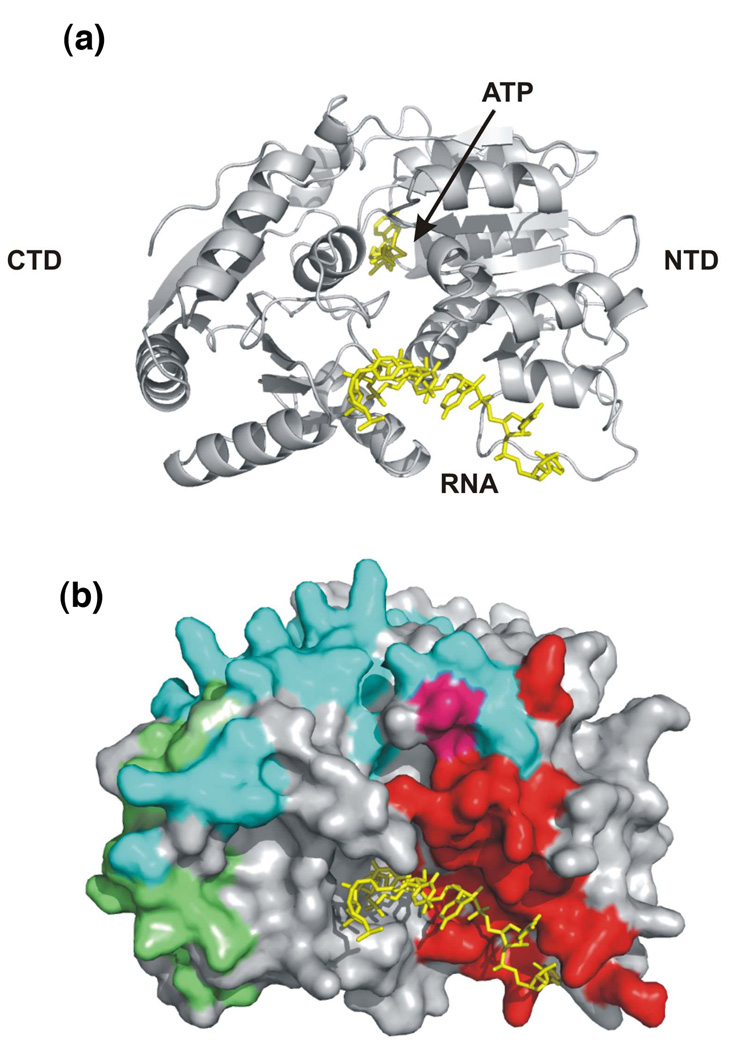
Figure 1.
Binding of different protein partners regulates the activity of DEAD-box RNA helicases. (a) Cartoon representation of the DEAD-BOX domains of DBP5 protein (grey): NTD and CTD are N-terminal and C-terminal RecA-like domains, respectively. The bound RNA and ATP are represented using yellow sticks (PDB ID: 3FHT; [14]). (b) Surface representation of the same molecule in the same orientation. The DBP5–NUP214 [14], Me31B–EDC3 [23] and eIF4AIII–MAGOH [21] (PDB IDs: 3FHC, 2WAX, 2HYI respectively)interaction surfaces are mapped on corresponding residues of DBP5 in red, green and cyan, respectively, with potentially overlapping residues mapped in a combination of the relevant colors. The figure highlights the existence of numerous, partially overlapping protein–protein (and protein–RNA)interaction surfaces of the DEAD-box helicase motif and the potential for a large number of alternative regulatory binding events. The inter-molecular surface was defined using the program InsightII (Accelerys), structural alignment of the different DEAD-box helicases was performed using the EBI DALI server [113], and the figures were created using the program Pymol (http://www.pymol.org).