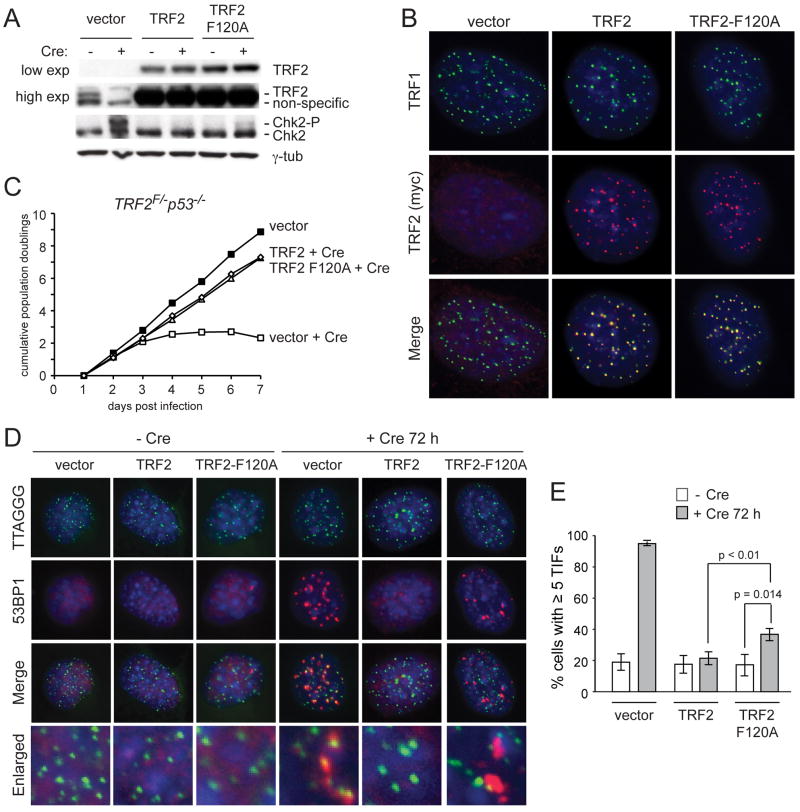
Figure 4. TRF2-F120A elicits a DNA damage response similar to Apollo deletion.
(A) Immunoblot for TRF2 and Chk2 in TRF2F/− p53−/− MEFs expressing the indicated TRF2 alleles without Cre and at 144 hours after Hit&Run Cre. (B) IF showing localization of TRF2 alleles in TRF2F/− p53−/− MEFs at 72h after Cre. TRF2 alleles are detected with the myc antibody 9B11. Telomeres are detected with the TRF1 antibody #644. (C) Growth curve showing cumulative population doublings after infection with Cre. Filled squares: vector (no Cre). Open squares: vector +Cre. Open circles: TRF2 +Cre. Open triangles: TRF2-F120A +Cre. (D) TIF assay on TRF2F/− p53−/− MEFs expressing the indicated TRF2 alleles to detect telomeric DNA damage signaling before (left) and after (right) deletion of the endogenous TRF2 with Cre. Telomeres are detected using a FISH probe (green). DNA damage sites are marked with 53BP1 (red). DNA is counterstained with DAPI (blue). (E) Quantification of TIF response as assayed in (D). TIFs were scored on the basis of co-localization of 53BP1 foci with 5 or more telomeres per cell. Values indicate the mean of 3 independent experiments (>100 nuclei per experiment) and SDs. p-values were determined based on paired student’s t-test.