Figure 3. Analysis of Timp3 and Rprm silencing in cultured mouse mammary tumor cell lines.
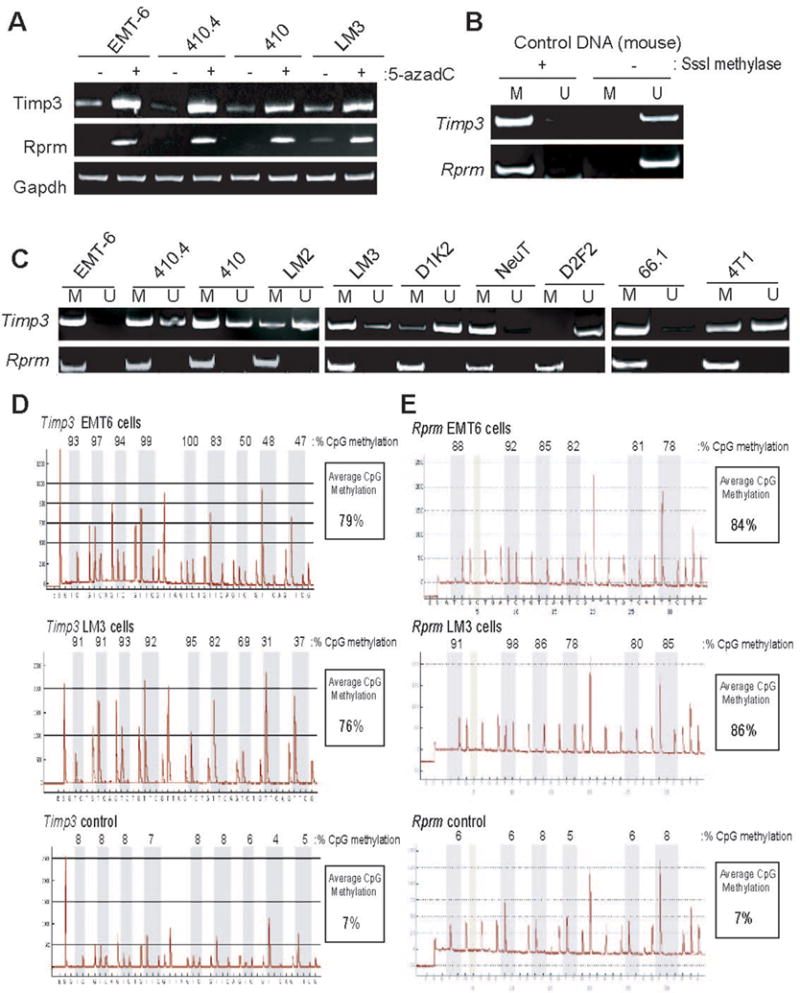
A Indicated mouse mammary tumor lines were either treated or not with a combination of 5-azadC and TSA. Subsequently, total RNA was harvested, cDNA synthesis performed, and PCR reactions conducted using primers specific for the Timp3 (top) or Rprm (middle) genes. Also, PCR reactions using primers specific for the Gapdh gene (bottom) were conducted to assure equivalent RNA abundance in each reaction. B. MSP primers were designed specific to the mouse Timp3 (top) and Rprm (bottom) genes. Primers designed to amplify methylated (M) or unmethylated (U) regions of the CpG island associated with each gene were tested in PCR reactions using mouse genomic DNA either incubated in the presence or absence of SssI methylase prior to bisulfite conversion. C. Genomic DNA harvested from mouse mammary tumor lines EMT6, 410.4, 410, LM3, LM2, NeuT, D2F2, 66.1, 4T1, D1K2 were bisulfite converted and used in MSP reactions with methylated-specific (M) or unmethylated-specific (U) primers. D. Pyrosequencing analysis was conducted on the Timp3 gene using bisulfite-modified genomic DNA from EMT6 and LM3 tumor lines, and mouse genomic DNA as a control. Shown are obtained pyrograms, measured methylation at CpG dinucleotides within the gene region analyzed, and the average CpG methylation within this region of the Timp3 gene. E. Pyrosequencing analysis of the Rprm gene was conducted on EMT6 and LM3 lines and mouse genomic DNA as outlined in D.
