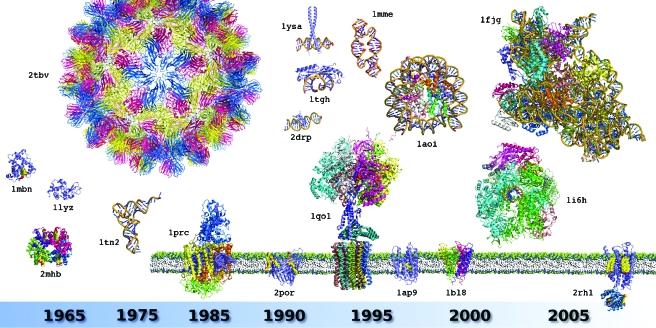
Figure 2. Milestone structures that recapitulate the history of biocrystallography.
The figure illustrates the diversity of 3D structures solved and the evolution of their complexity since the birth of the field in the late 1950s. A star ( *) indicates crystal structures linked to a Nobel Prize award. The proposed selection is displayed in chronological rank and includes: sperm whale myoglobin (PDB identifier: 1mbn*), horse hemoglobin (2mhb*), hen egg white lysozyme (1lyz), Saccharomyces cerevisiae tRNAPhe (1tn2), icosahedral Tomato Bushy Stunt Virus (2tbv), Rhodopseudomonas viridis photosynthetic reaction center (1prc*), Rhodobacter capsulatus porin (2por), human TATA binding protein in complex with TATA box DNA (1tgh), S. cerevisiae GCN4 leucine zipper (1ysa), Drosophila melanogaster Tramtrack zinc finger domain complexed with its DNA target (2drp), bovine ATP synthase (1qo1*), synthetic construct of a hammerhead ribozyme (1mme), Xenopus laevis nucleosome with synthetic DNA construct (1aoi), Halobacterium salinarum bacteriorhodopsin (1ap9), Streptomyces lividans K+ channel (1bl8*), S. cerevisiae RNA polymerase II (1i6h*), Thermus thermophilus 30S ribosomal subunit (1fjg*), and a human G Protein Coupled Receptor or GPCR (2rh1). Ligands, cofactors, and additives are shown in CPK form. For details and other milestones, see Supplementary Material Table S1. All structures are displayed at the same scale using PyMol (Delano Scientific—http://www.pymol.org). Membrane proteins are shown with their trans membrane region emphasized in a schematized membrane (notice the lysozyme module fused to the intracellular part of the GPCR structure, see text for details).