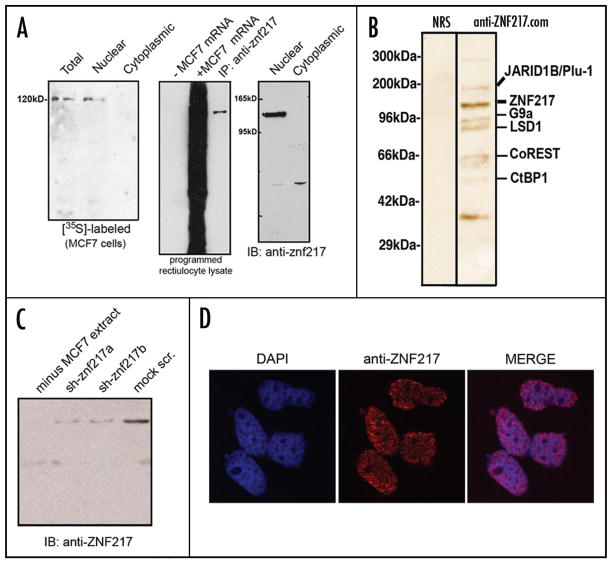
Figure 1.
Nuclear ZNF217 binding proteins identified by affinity chromatography. (A) Fidelity of ZNF217 antisera. Western blotting and immunoprecipitation illustrates the specificity of the anti-ZNF217 antibody used in this study, which stains ZNF217 as a single band. Using [35S]-labeled cellular extract fractions pulse-labeled with [35S]-methionine/cysteine was used to detect ZNF217 by immunopreciptation in nuclear and cytosolic fractions. (B) Affinity chromatographic purification of ZNF217 binding proteins was performed capturing endogenous ZNF217 from nuclear extract with anti-ZNF217 antibodies (bound to the column) under non-denaturing conditions. Shown is an SDS gel that was silver-stained. Individual protein bands were extracted, subjected to tryptic digestion, and analyzed with MALDI-TOF mass spectrometry. The molecular weight of all proteins identified by this method was consistent with their known sizes. (C) RNA interference of ZNF217 by two specific siRNAs against ZNF217 in MCF7 cells was used to determine the presence of ZNF217 following transfections of siRNAs. (D) Immunocytochemical staining of MCF7 cells using ZNF217 antisera followed by secondary labeling with anti rabbit IgG conjugated with Texas Red (Thermo Scientific/Pierce). Fixed cells were counter-stained with DAPI and DAPI labeling was used to merge contrasting images to confirm co-localization.