Abstract
Interferon-γ (IFN-γ) stimulation triggers tyrosine-phosphorylation of transcription factor STAT1 at position 701, which is associated with the switching from carrier-independent nucleocytoplasmic shuttling to carrier-mediated nuclear import. Unlike most substrates that carry a classical Nuclear Localization Signal (cNLS) and bind to importin α1, STAT1 possesses a non-classical NLS recognized by the isoform importin α5. In the present study, we have analyzed the mechanisms by which importin α5 binds phosphorylated STAT1 (pSTAT1). We found that a homodimer of pSTAT1 is recognized by one equivalent of importin α5 with Kd = 191 ± 20 nM. Whereas tyrosine-phosphorylation at position 701 is essential to assemble a pSTAT1:importin α5 complex, the phosphate moiety is not a direct binding determinant for importin α5. In contrast to classical NLS substrates, pSTAT1 binding to importin α5 is not displaced by the N-terminal importin β binding (IBB)-domain, and requires the importin α5 C-terminal acidic tail (505-EEDD-508). A local unfolding of importin α5 ARM repeat 10 accompanies high affinity binding to pSTAT1. This unfolding is mediated by a single conserved tyrosine at position 476 of importin α5, which is inserted in between Armadillo (ARM) repeat 10 helices H1-H2-H3, thereby preventing the intramolecular helical stacking essential to stabilize the folding conformation of ARM 10. Introducing a glycine at this position, as in importin α1, disrupts high-affinity binding to pSTAT1, suggesting pSTAT1 recognition is dependent on the intrinsic flexibility of ARM 10. Using the quantitative stoichiometry and binding data presented in this paper together with mutational information available in the literature, we propose importin α5 binds in between two STAT1 monomers, with two major binding determinants in the SH2- and DNA-binding domains. In vitro this model is supported by the observation that a 38mer DNA oligonucleotide containing two tandem cfosM67-promoters can displace importin α5 from pSTAT1, suggesting a possible role for DNA in releasing activated STAT1 in the cell nucleus.
Keywords: Nuclear Import, STAT1, importin α5, importin β, NLS, non-classical NLS, phospho-tyrosine, molecular recognition
INTRODUCTION
The JAK/STAT pathway, a key player in the host immune response, transmits a cytokine signal from the extra-cellular space to the nucleus of the host cell to drive the transcription of genes responsible for host defense 1. The products from seven different STAT genes are expressed in mammals, which share a fundamentally conserved topology consisting of three independent structural subunits 1. The N-terminal helical domain, roughly 130 amino acids in length, promotes dimerization of the unphosphorylated STATs and polymerization of phosphorylated dimers on DNA 2; 3. This is followed by a large STAT core of approximately 600 residues, containing a coiled-coil domain, a DNA binding domain and an SH2-domain. Before cytokine stimulation of cells, STAT1 is a dimer that adopts anti-parallel conformation involving N-domain interactions 4. Structural evidence indicates that such dimers are recruited to cytokine receptors for activation by phosphorylation of a C-terminal tyrosine residue 5; 6. Upon activation, an additional homodimer conformation emerges where the phosphotyrosine (pTyr) of one protomer reaches into the SH2 domain of the other 7. SH2-pTyr-mediated dimers, also called parallel dimers, possess DNA-binding activity and evoke the transcriptional responses to cytokines. While unphosphorylated STAT1 is a nucleocytoplasmic shuttling protein that efficiently enters the nucleus independent of carrier molecules, nuclear import of activated STATs requires importins. Moreover, activated STAT1 is barred from nuclear export. Although pSTAT1 is efficiently dephosphorylated in the nucleus by phosphatase TC45 (t½ ~20 min) 8, protracted nuclear export becomes apparent as nuclear accumulation of STAT1 within minutes of cytokine stimulation 9. These mechanisms ensure nuclear retention of the active transcription factor, while linking the activity of cytokine receptors to their effector functions in the nucleus. Vaccinia virus and other members of the Poxviridae family of double stranded DNA viruses 10 encode the dual specificity phosphatase VH1 that specifically dephoshorylates activated STAT1 11; 12. In contrast to TC45, VH1 is thought to dephosphorylate STAT1 in the cytoplasm, thus preventing the expression of antiviral genes 12.
The carrier-dependent passage of proteins through the nuclear pore complex (NPC) is an active, signal-mediated process that requires soluble transport factors of the importin β-superfamily (also known as β-karyopherins) and the small GTPase Ran 13; 14; 15; 16; 17. Import complexes form in the cytoplasm upon recognition of a nuclear localization signal (NLS) in import cargos by β-karyopherins. This interaction can be direct, or mediated by transport adaptors such as importin α and snurportin 18. There are six human importin α isoforms in humans that show greater than 60% sequence similarity 19 and fall into three phylogenetically distinct groups, the α1s, α2s and α3s 19. All importin αs are made up of 10 stacked Armadillo (ARM) repeats 20, each formed by three α-helices 19. Different importin α isoforms have striking differences in substrate recognition, which enhances the specificity for nuclear import of diverse import cargos, but also share the ability to bind and import classical NLS substrates 21. Despite the presence of six importin α isoforms, the majority of import cargos contain a classical SV40-like NLS that is recognized by importin α1 13. Structural work has shown that the basic side chains of an NLS occupy a shallow groove within the ARM repeats 1-4 of importin α, which is known as the major binding site, as well as a minor binding site between ARM repeats 4-8. Five points of contact between NLS and importin α (usually referred as positions P1-P5) have been identified from the analysis of several importin α/NLS complexes 22; 23; 24; 25; 26; 27. Structural and mutational data have demonstrated the crucial role played by the NLS basic lysine at the P2 site, which contacts the Trp/Asn pair between importin α1 ARM repeats 3 and 4 28, 24; 29. Interestingly, certain animal isoforms are very specific for non-classical cargos. For instance, importin α5 (also known as NPI-1 30) is involved in the nuclear import of dimeric phosphorylated STAT1 31, and influenza virus polymerase PB2 32. The crystal structure of importin α5 was recently determined in complex with the C-terminal domain of influenza virus RNA polymerase PB2 32. In this structure, the C-terminal folded core of PB2 (res. 686-741) makes extensive interactions with the C-terminus of importin α5, where ARM repeat 10 deviates from its canonical structure to wrap around the polymerase subunit 32. In addition, an extended N-terminal moiety of PB2 (res. 749-757) extends inside the major NLS-binding site (ARM repeats 1-4) to adopt a stretched conformation similar to that seen for the SV40-NLS 24. On overall, the recognition of PB2 by importin α5 is more extended and complex than that of classical NLS-peptides bound to importin α1 and requires the C-terminal domain of importin α5. Despite differences in cargo specificity and recognition, all importin α isoforms bind the receptor importin β via an N-terminal importin β binding (IBB) domain. Import complexes formed in the cytoplasm move through the NPC in a process that likely involves multiple rounds of interactions with phenylalanine-glycine nucleoporins (FG-nups). Ran is naturally compartmentalized in eukaryotic cells, with a nuclear RanGTP pool, while cytoplasmic Ran is predominately in the RanGDP form 13; 15. RanGTP facilitates translocation through the NPC by releasing import complexes from high affinity binding sites in nucleoporins and disassembling import complexes in the cell nucleus 13; 15. GTP hydrolysis by the small GTPase Ran is thought to impose directionality of transport through the NPC 33.
Mounting evidence has suggested the nuclear import of activated STAT1 is functionally different from the import of classical NLS (cNLS)-bearing cargos 31. Activated STAT1 lacks a cNLS in its primary sequence, and is imported by importin β in complex with the isoform importin α5/NPI-1 34. Although importin α5 can also mediate nuclear import of cNLS-cargos in permeabilized cells 21, cNLS peptides fail to compete off pSTAT1 from importin α5, suggesting the binding site of pSTAT1 in importin α5 is distinct from that of classical NLS-cargos34. Instead, STAT1 contains a non-classical NLS in the DNA binding domain that is exposed only upon phosphorylation-induced dimerization, and therefore, referred to as a dimer-specific NLS (dsNLS) 35; 36; 37; 38. Unlike the cNLS, the STAT1-dsNLS is a non-transferable nuclear targeting sequence, functional only in the context of activated STAT1 35. Furthermore, recognition of the pSTAT1-dsNLS by importin α5 requires the intact STAT1 N-terminal domains, as dimeric phosphorylated STAT1 core alone has no detectable affinity for the transport machinery 39.
In the present study, we dissect the mechanisms by which importin α5 recognizes phosphorylated STAT1. We found that the unfolding of importin α5 C-terminal ARM 10 is essential for high-affinity recognition of pSTAT1. This event is mediated by a conserved ‘Tyr-finger’ at position 476 of importin α5 that prevents ARM 10 from adopting a closed conformation, as seen in importin α1 28. The binding determinant for pSTAT1 in ARM 10 can be transferred in trans to importin α1, therefore suggesting that pSTAT1 uses determinants also present in the cNLS binding groove in addition to a unique architecture of the C-terminal ARM 10. Using the data presented in this study and the information reported in the literature, we propose a structural model of pSTAT1 binding to importin α5.
RESULTS
A single importin α5 molecule binds a dimer of pSTAT1
As a first attempt to characterize the recognition of pSTAT1 by importin α5, we sought to determine the stoichiometry of the pSTAT1:importinα5 import complex in solution, using homogeneously purified proteins. We expressed full length STAT1 in E. coli and after purification by metal chelating chromatography, we phosphorylated STAT1 in vitro using purified epidermal growth factor (EGF) receptor immobilized on protein A-agarose beads 2. To ensure that STAT1 was biochemically homogeneous, pSTAT1 was separated from the unphosphorylated STAT1 (uSTAT1) population by heparin chromatography, followed by gel-filtration chromatography. This purification yielded homogeneously pure pSTAT1, which is fully active in DNA binding (data not shown). To define the stoichiometry of the STAT1 import complex, we subjected purified unphosphorylated and phosphorylated STAT1, importin α5, and mixtures thereof at different molar ratios to analytical gel-filtration chromatography. At physiological salt concentration and pH, uSTAT1, pSTAT1, and importin α5 migrated as mono-disperse species eluting after ~11 ml, 9.5 ml, and 13.5 ml, respectively (Fig. 1A). Molecular weight calibration standards indicated for importin α5 an apparent molecular weight of ~100 kDa (monomer 62 kDa). uSTAT1 and pSTAT1 migrated as ~400 kDa and 600 kDa species, respectively, thus both appeared significantly larger than expected from the theoretical M.W. for a dimer of ~180 kDa (Fig. 1A). When pSTAT1 was mixed with an equimolar amount of importin α5, two peaks appeared, the second of which completely overlapped the peak position of free importin α5 (Fig. 1A). SDS-PAGE followed by Western blot analyses showed that it contained predominantly importin α5 with little STAT1 (Fig. 1B, top panel). The first peak with an elution volume between uSTAT1 and pSTAT1 (Fig. 1A) contained not only the bulk of pSTAT1, but also substantial amounts of importin α5 (Fig. 1B, top panel), indicating complex formation between the two proteins. Next, importin was added in excess over pSTAT1 (2:1), which resulted solely in the increased elution of free importin α5 (Fig. 1A, B, middle panel). The inverse molar ratio (1:2), namely one equivalent of importin α5 mixed with pSTAT1 dimer, resulted in the disappearance of free importin α5 (Fig. 1A, B, bottom panel). We thus concluded that one molecule of importin α5 was binding one pSTAT1 dimer. Of note, similar observations were not made when importin α5 was mixed with uSTAT1, which regardless of their mixing ratio eluted as independent molecular species (not shown), confirming that complex formation required STAT1 activation.
Figure 1. Stoichiometry of STAT1 nuclear import complex.
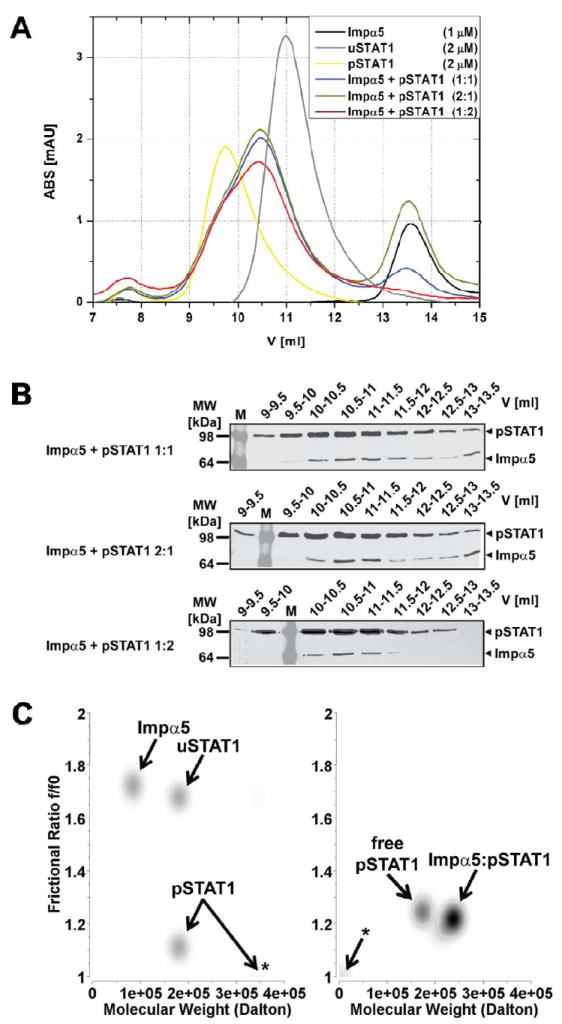
(A) Gel-filtration analysis of purified Strep-tagged unphosphorylated STAT1 (uSTAT1) and phosphorylated-Tyr701-STAT1 (pSTAT1), importin α5 (Impα5) and varying molar mixtures of pSTAT1 and Impα5. Free pSTAT1 elutes at 9.7 ml (presumably in a dimer-tetramer equilibrium) and free uSTAT1 at 11 ml (presumably as a dimer); the pSTAT1:importin α5 complexes formed at different molar ratios of pSTAT1 and Impα5 elute at identical elution volumes of 10.5 ml. Free Impα5, and excess Impα5 not bound in the pSTAT1:importin α5 complex elutes at 13.5 ml. (B) The indicated elution fractions obtained from the gel-filtration analyses of pSTAT1:importin α5 complexes shown in (A) were analyzed by Western Blot with anti-Strep antibody after 10% SDS-PAGE. (C) Sedimentation velocity analyses of uSTAT1, pSTAT1, importin α5, and complexes thereof. Graphical representation of molecular species detected by genetic algorithm–Monte Carlo analysis with molecular weights (M.W.) and frictional ratio in pseudo-3D plots. Left panel, Impα5, uSTAT1, and pSTAT1 were analyzed separately. Note an additional high molecular weight peak for pSTAT1 consistent with tetramerization (asterisk). Right panel, Impα:pSTAT1 was mixed and gel-filtrated. The peak fraction containing Impα:pSTAT1 complex was used in the ultracentrifugation experiment. Two molecular species with molecular weights of ~180 kDa and ~245 kDa were present. The latter was assigned to represent pSTAT1:importin α5 complex, whereas the former represents free pSTAT1 dimers. Asterisk, co-sedimenting salt in the reference cell.
Additionally, we used analytical ultracentrifugation to independently examine the molecular weights, shapes, and associations of STAT1 and importin α5. Sedimentation velocity experiments with the isolated importin α5 demonstrated its M.W. to be 90 kDa, which suggested monomer-to-dimer transition of this molecule (Fig. 1C, left panel). For uSTAT1 and pSTAT1 this experimental approach showed identical molecular weights of ~180 kDa, which confirmed stable dimerization regardless of the phosphorylation state of STAT1. However, the dimers differed starkly in their shapes, as indicated by the frictional ratio of 1.7 for uSTAT1 and 1.1 for pSTAT1 (Fig. 1C, left panel). Moreover, pSTAT1 appeared to form additional higher molecular weight-complexes, probably tetramers, as observed previously 4. These remarkable differences are likely to explain the different elution profiles of uSTAT1 and pSTAT1 shown in Fig. 1A. Next, pSTAT1:importin α5-complex was isolated by preparative gel-filtration and subjected to sedimentation velocity analyses (Fig. 1C, right panel). The gel-filtration peak contained two molecular species with M.W. of ~180 kDa and ~240 kDa, respectively, and identical frictional ratios of ~1.2. We thus concluded that the ~180 kDa species represents excess pSTAT1 dimer, which cannot reliably be removed from the pSTAT1-importin-complex by gel-filtration (Fig. 1A). Since we did not detect traces of free importin α5 in this peak, we concluded that the free pSTAT1 did not result from dissociation of pSTAT1:importin during the centrifugation, but was genuinely free pSTAT1 that had remained unbound; probably because some importin α5 was incompetent for binding. The M.W. of ~240 kDa and the frictional ratio of the second molecular species suggested that it represents pSTAT1 dimers (M.W. ~180 kDa) bound to importin α5 monomer (M.W. 62 kDa). Thus, the stoichiometry of the pSTAT1:importin α5 import complex determined by analytical ultracentrifugation is consistent with a single adaptor molecule per dimer of pSTAT1, and thus agrees with the gel-filtration data.
Phospho-Tyr701 in STAT1 is not a binding determinant for importin α5
STAT1 is phosphorylated at Tyr701, and phosphorylation is essential for energy-dependent nuclear import 40. However, it is unknown whether the phosphate moiety at position 701 serves as a direct binding determinant for the selective recognition by importin α5. This is a plausible hypothesis, as phosphorylation of Ser385 in the NLS Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA-1) is known to up-regulate binding to importin α5 and EBNA-1 nuclear transport 41. We used isothermal titration calorimetry (ITC) to determine the specificity of importin α5 for phospo-Tyr701. As a proof of concept that ITC can be used to measure the contribution of a single phosphate moiety in a functional NLS, phosphorylated EBNA-1 NLS peptide (379-KRPRSPpSS-386) was injected into an ITC cell containing 50 μM importin α5 and the enthalpy of binding was carefully monitored (Fig. 2A). In a control experiment, unphosphorylated EBNA-1 NLS peptide (379-KRPRSPSS-386) was injected against importin α5 under identical experiment conditions (Fig. 2B). In agreement with the published data 41, we observed that the enthalpy released upon binding of the EBNA-1 NLS to importin α5 was 20-fold greater when Ser385 is phosphorylated (Fig. 2, compare panel A to B). The Kd estimated for the pNLS is ~3 μM as compared to ~60 μM for the unphosphorylated NLS. This strongly supports the idea that the phosphate moiety within EBNA-1 NLS functions as a direct binding determinant for importin α5. To determine whether importin α5 recognizes the STAT1 phospho-Tyr701 directly, a peptide spanning STAT1 region 695-708 containing a phspho-Tyr701 was synthesized and probed for binding to importin α5 by ITC, under identical experimental conditions (as in panel A and B). Interestingly, no interaction between the STAT1-peptide and importin α5 was observed, even at high micromolar concentration (Fig. 2C). Thus, importin α5 can bind the Ser-phosphorylated NLS of EBNA-1 with high affinity, but STAT1 phospho-Tyr701 is not a direct binding determinant for importin α5. The phosphorylation of STAT1 is likely to induce a conformational change 4 that exposes a STAT1-dsNLS for binding to importin α5.
Figure 2. ITC analysis of the interaction of phosphorylated NLS peptides with importin α5.
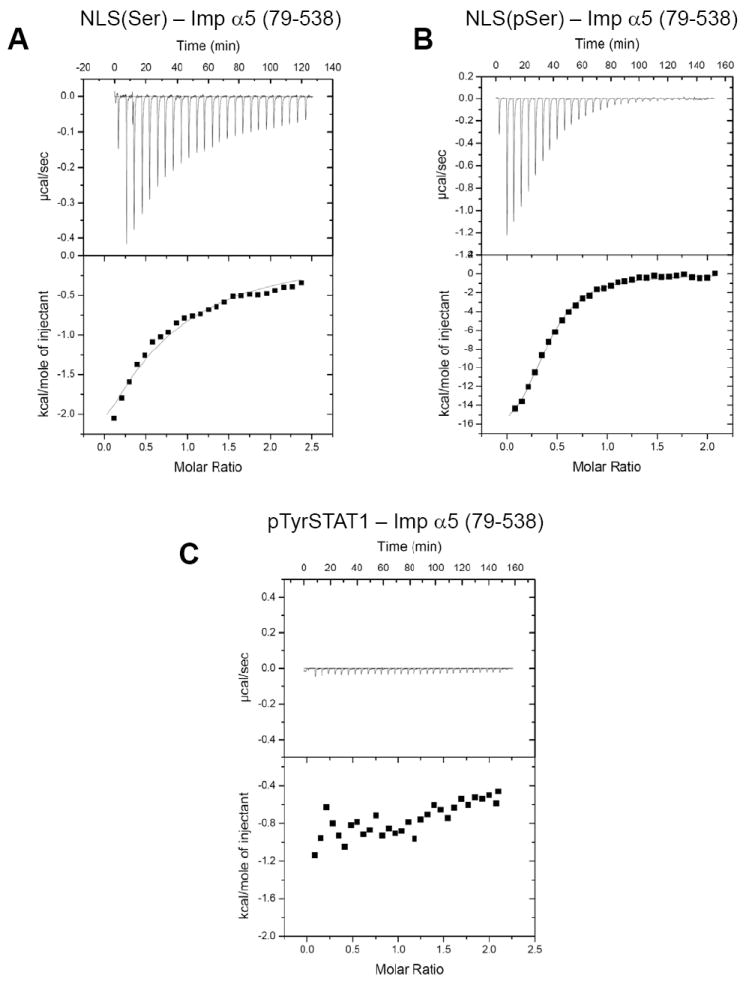
(A) Injection of unphosphorylated EBNA-1 NLS peptide (379-KRPRSPSS-386) into a cell containing importin α5. (B) Injection of EBNA-1 phospho-Ser385NLS peptide (379-KRPRSPpSS-386) into importin α5. Notice that the heat released in panel B is three times greater than in panel A. (C) Injection of pTyr701 STAT1 peptide spanning residues 695-708 into a cell containing importin α5. No significant heat release is observed. In (A-C), the raw data are in the top panel, and the integrated enthalpy plotted as a function of the NLS:importin α5 molar ratio is shown in the bottom panel. The dissociation constants measured from the ITC data for the interaction of EBNA-1 NLS peptide and phospho-Ser385pNLS peptide with importin α5 are Kd= 60 ± 14 μM and Kd= 3 ± 5 μM, respectively.
Importin α5 binds activated dimeric STAT1 with nanomolar affinity
We then used surface plasmon resonance (SPR) to obtain a quantitative kinetic description of the binding affinity of importin α5 for activated pSTAT1. Increasing concentrations of pSTAT1 was flowed into the cell containing GST-importin α5 immobilized on a biosensor surface. Each interaction was measured using five different concentrations of analyte (pSTAT1) and each concentration was repeated four times. pSTAT1 binds full length importin α5 with a Kd ~191 ± 20 nM (Fig. 3A, Table I), and the rate of pSTAT1 association to and dissociation from importin α5 measured by SPR are kon = 9.88 e4± 1.0 e4 (M-1 s-1) and koff = 18.3 e-3± 0.5 e-4 (s-1), respectively (Table I). No significant binding to importin α5 was observed using either uSTAT1, or pSTAT1 core (res. 132-710), which lacks the N-terminal dimerization domain (Table I). Likewise, simultaneous addition of STAT1 N-terminal domain and pSTAT1 core did not restore binding to importin α5 (Table I). This suggests that STAT1 N-terminal domain must be fused to the main pSTAT1 core to allow for high affinity binding to importin α5. However, the point mutations Phe77Ala and Leu78Ala in pSTAT1 that disrupt N-terminal dimerization, but leave pSTAT1 dimeric 42 yielded a comparable binding affinity for importin α5 as wild type pSTAT1 (Kd ~ 112 ± 22 versus 191 ± 20 nM) (Table I). This indicates STAT1 N-terminal domains, and not their homotypic interaction, are essential for importin α5 binding. Interestingly, removal of importin α5 IBB-domain (res. 1-78) did not significantly affect the binding affinity for pSTAT1 (Kd ~153 ± 16 nM) (Fig. 3B, Table I), which suggests that the IBB-domain does not auto-inhibit pSTAT1 binding, as shown for the cNLS-bearing cargos binding to importin α1 28; 43; 44.
Figure 3. Surface plasmon resonance (SPR) analysis of the interaction of importin α5 with pSTAT1.
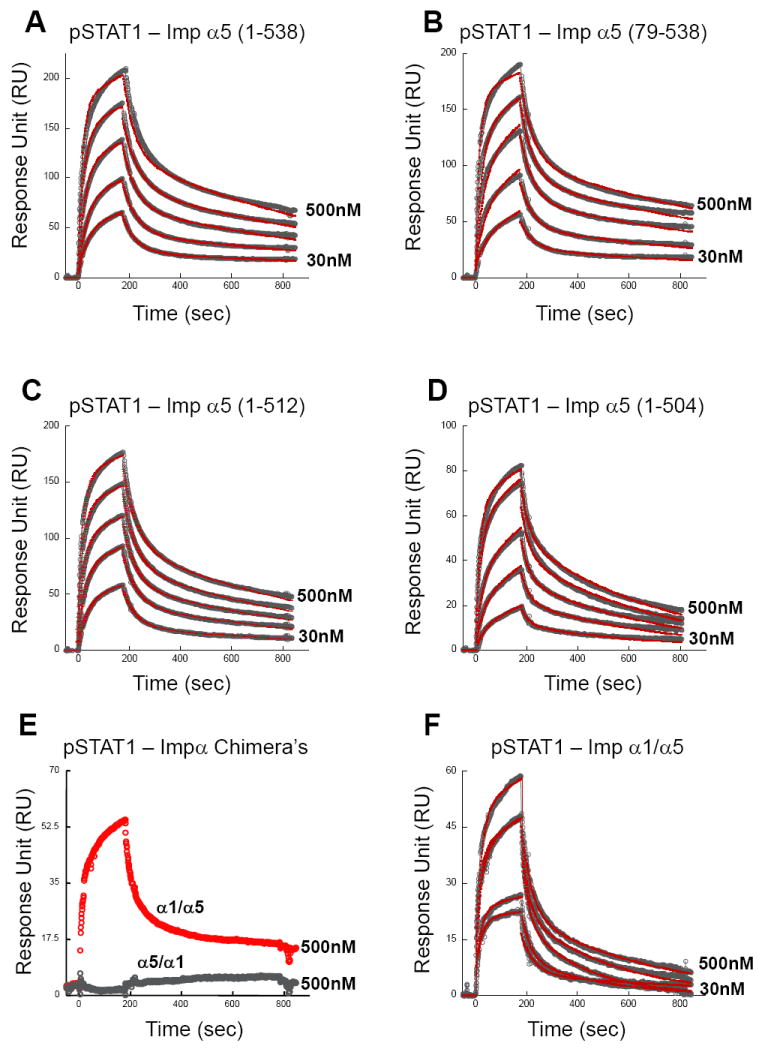
(A) GST-importin α5 (res. 1-538), (B) GST-ΔIBB-importin α5 (res. 79-538), (C) GST-importin α5 (res. 1-512), (D) GST-importin α5 (res. 1-504), (E) GST-importin α1/α5 (res. 1-446 of α1/449-538 of α5) and (F) GST-importin α5/α1 (res. 1-449 of α5/447-529 of α1) were captured onto a sensor chip using an immobilized anti-GST antibody. Purified pSTAT1 (the analyte) was flown into the cell between concentrations of 30, 62.5, 125, 250 and 500 nM, at a flow rate of 30 μl/min. In each sensogram, raw data are shown as open circles and fit curves as red lines for five different concentrations of analyte. In panel (E), only raw data for one representative concentration of pSTAT1 is shown, corresponding to 500 nM. The sensogram for GST-importin α1/α5 binding to pSTAT1 (in red) is also included to better show the complete loss of GST-importin α5/α1 binding to pSTAT1 (in gray). A complete list of kinetic parameters is in Table I.
Table I.
A complete list of all kinetic parameters measured by SPR to study the interaction of importin α5 with pSTAT1. kon and koff are the association and dissociation rate constants; Kd is the dissociation constant. χ2 is the chi-square test to compare how observed data correlate with fitting models used to determine Kd values.
| GST-Imp α | Analyte | Kd* (nM) | kon (M-1 s-1) | koff (s-1) | χ2 |
|---|---|---|---|---|---|
| α5 (1-538) | pSTAT1 (1-712) | 191 ± 20 | 9.88e4 | .0189 | .485 |
| α5 (1-538) | uSTAT1 (1-712) | N/D | N/D | N/D | N/D |
| α5 (1-538) | pSTAT1 core (132-712) | N/D | N/D | N/D | N/D |
| α5 (1-538) | pSTAT1 core (132-712) + ND (1-132) | N/D | N/D | N/D | N/D |
| α5 (1-538) | pSTAT1 (1-712) F77A / L78A | 112 ± 22 | 9.81e4 | .0113 | 1.22 |
| α5 (79-538) | pSTAT1 (1-712) | 153 ± 16 | 1.44e5 | .0218 | .603 |
| α5 (1-512) | pSTAT1 (1-712) | 166 ± 17 | 1.24e5 | .0204 | 1.12 |
| α5 (1-504) | pSTAT1 (1-712) | 247 ± 27 | 9.83e4 | .0243 | .84 |
| α1 / α5 | pSTAT1 (1-712) | 542 ± 36 | 5.39e5 | .0284 | .98 |
| α5 / α1 | pSTAT1 (1-712) | N/D | N/D | N/D | N/D |
| α5 (1-538) Tyr476Gly | pSTAT1 (1-712) | 4,300 ± 140 | 6.66e4 | .289 | 2.9 |
Dissociation constant Kd = (koff/kon)
To characterize the role of importin α5 C-terminus in binding to pSTAT1, we first deleted the last 25 residues of importin α5 spanning region 512-538. By SPR, this construct displayed wild type binding affinity for pSTAT1 (Kd ~166 ± 17nM) (Fig. 3C, Table I). In a separate construct, we further removed importin α5 residues 505-510, which contain the acidic ‘tail’ 505Glu-Glu-Asp-Asp510, previously implicated in binding to the export receptor CAS 45. Notably, importin α5 (1-504) had slightly reduced binding affinity for pSTAT1 (Kd ~247 ± 27 nM) (Fig. 3D, Table I) as compared to the wild type protein, primarily due to a faster rate of pSTAT1 dissociation (koff = 24.3 e-3± 0.5 e-4 versus 18.3 e-3± 0.5 e-4 s-1). This suggests importin α5 acidic tail is likely used to stabilize pSTAT1 and prevent its dissociation. Unfortunately, deletions upstream of residue 504 yielded unstable importin α5 constructs, with a high tendency to dimerize and aggregate, which prevented quantitative analysis by SPR.
To extend our quantitative binding analysis to importin α5 residues upstream of position 504, we generated a chimeric adaptor protein where importin α5 residues 449-538 were replaced by the equivalent residues of importin α1 (447-529). This chimera, which we named importin α5/α1, was completely inactive in pSTAT1 binding (Fig. 3E, Table I), thus supporting previous deletion studies by pull-down assay 34; 46. Conversely, a chimera of importin α1 containing residues 449-538 of importin α5 showed saturable binding to pSTAT1 with Kd ~542 ± 30nM (Fig. 3F, Table I), thereby demonstrating that the binding determinants in importin α5 ARM 10 responsible for high affinity binding to pSTAT1 can be transferred in trans to another isoform of the transport adaptor. Although the affinity of importin α1/α5 for pSTAT1 is ~2.5 fold lower than that measured for wild type importin α5 (Kd ~542 versus 190 nM), the importin α1/α5 chimera has an artificial interface between ARM 9 and 10, which may be responsible for the observed drop in affinity when compared to wild type importin α5. Thus, kinetic analysis by SPR demonstrates that importin α5 binds the pSTAT1 with nanomolar affinity. This interaction is largely mediated by importin α5 ARM 10 and is independent of both the IBB-domain and C-terminal tail (res. 512-538). Also, the dimerization of pSTAT1 N-terminal domains is not a critical determinant for binding to importin α5.
Tyrosine 476 in ARM 10 of importin α5 is critical for pSTAT1 recognition
To further understand the molecular specificity of importin α5 for pSTAT1, we focused on the structural differences between this isoform of importin α and the classical adaptor α1. The structure of importin α5 was recently determined in complex with the C-terminal domain of influenza virus RNA polymerase PB2 32. In this structure, the binding interface between adaptor and cargo spans the entire length of importin α5, where it occupies both major and minor NLS recognition sites, together with ARM repeat 10 32. Comparison of this structure with the mammalian importin α1 25; 28 reveals a dramatic unfolding of ARM 10 in response to cargo binding (Fig. 4A). However, in importin α1, ARM 10 is folded as a conventional ARM repeat, with helices H1-H2-H3 stacked onto each other to generate a flat surface, in importin α5, ARM 10 helices H2 and H3 swing away by ~40° and 90°, respectively from the corresponding position in α1 (Fig. 4B). Unfolding ARM 10 results in a molecular embracing of polymerase PB2, which binds importin α5 with high affinity, but, like pSTAT1, has no significant affinity for importin α1 32. To determine the structural determinants that allow importin α5 ARM 10 to unfold upon substrate binding, we analyzed the primary sequence of the six human 19 importin α isoforms (Fig. 4C). Interestingly, we found that a conserved Tyr residue at position 476 of importin α5, α6 and α7 (the NPI-1-like importin αs) is always replaced by a Gly in all other isoforms. This is quite intriguing as, like importin α5, importin α7 was also shown to bind STAT1 47. Structurally, Tyr476 is located at the end of the first helix of ARM 10 (helix H1) where it projects its bulky aromatic side chains into the interface of ARM 10 helices H1-H2-H3 (Fig. 4B). This ‘Tyr-finger’ is likely to destabilize the stacking of the three ARM helices, thereby allowing for local unfolding of ARM 10 helix H3. In contrast, in other importin α isoforms where Tyr476 is replaced by a Gly (Gly468 in the universal importin α1) (Fig. 4C), ARM 10 helices H1, H2 and H3 pack onto each other via hydrophobic interactions to generate a flat and rigid structure, characteristic of ARM repeats.
Figure 4. A conserved tyrosine at position 476 of importin α5 allows unfolding of ARM 10.
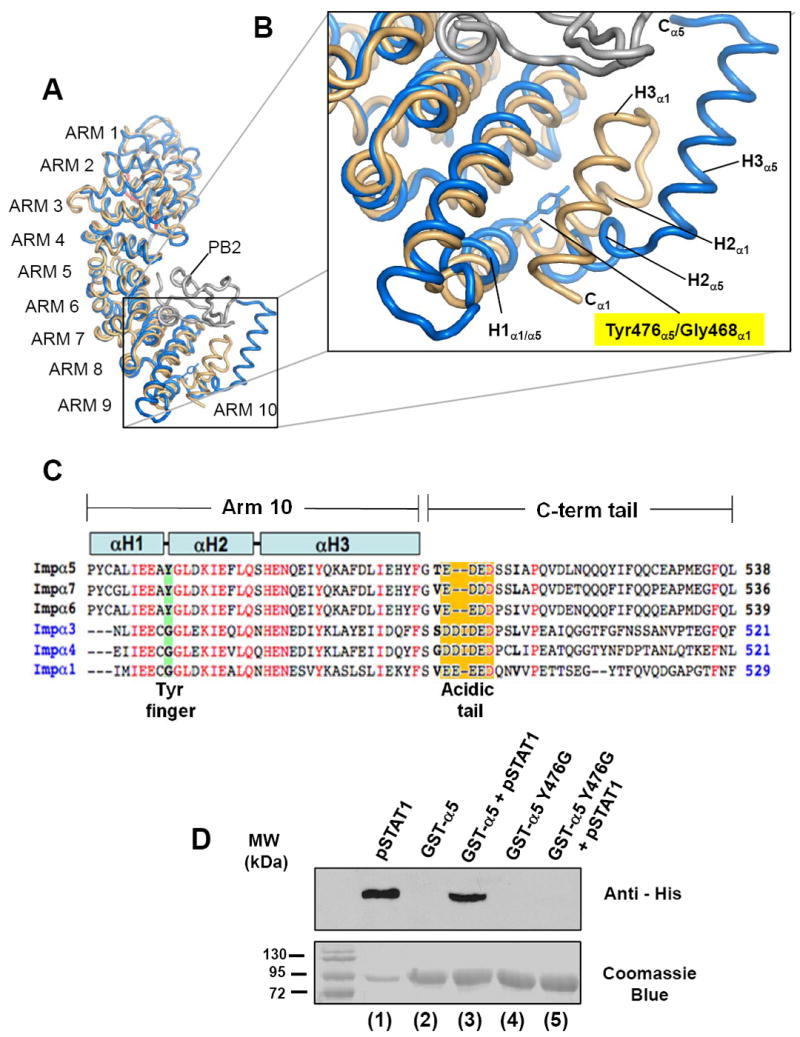
(A) Superimposition of importin α5 bound to the C-terminal domain of influenza virus polymerase PB2 (pdb # 2JDQ) 32 and importin α1 (pdb # 1Y2A) 26. Importin α5 and α1 are colored in beige and blue, respectively, while influenza PB2 is in gray. (B) Blow-up of ARM 10 with Tyr476 and Gly468 in importin α5 and α1, respectively shown as stick-and-balls. (C) Sequence alignment of human importin αs C-terminal residues 467-538. Highlighted in green is position 476, which is Tyr in the NPI-1 subfamily (α5, α6 and α7) and Gly in other importin α’s. Highlighted in orange is the poly-acidic tail, that in importin α5 spans residues 504-508. In the alignment, identical residues are colored in red. (D) Pull-down assay. Top panel is a Western blot of fractions probed with a monoclonal anti-His tag antibody. Bottom panel is the SDS-gel used for blotting. Lane 1 is a control of his-pSTAT1, which is efficiently recognized by the anti-His antibody. Lane 3 and 5 contain pSTAT1 pulled down by GST-importin α5 (lane 3) and GST-importin α5 (Tyr476Gly)-importin α5 (lane 5), respectively. The Tyr476Gly mutant has completely lost binding to pSTAT1. Lanes 2 and 4 are controls to show that neither wild type GST-importin α5 nor GST-importin α5 (Tyr476Gly) react with anti-His antibody.
Based on the structural evidences presented in Fig. 4A-B, we hypothesized that the unfolding of ARM 10 helix H3 in importin α5 is essential for high affinity binding of non-classical cargos like influenza PB2 and pSTAT1. To test this hypothesis, we replaced Tyr476 in importin α5 to Gly, which mimics the ARM 10 found in importin α1. Interestingly, importin α5 Tyr476Gly had no detectable binding affinity to pSTAT1 by pull-down assay (Fig. 4D, lane 5). By SPR, pSTAT1 displayed dramatically high dissociation rate from importin α5 Tyr476Gly with a koff ~ 289.0 s-1 as compared to koff 18.3 e-3 s-1 measured for wild type importin α5, and an overall binding affinity that could be estimated to be in the micromolar range (Table I). In a converse experiment, a Tyr residue was introduced at the equivalent position of importin α1 to determine if this mutation would result in a gain of function. However, by pull-down assay importin α1 (Gly468Tyr) failed to bind pSTAT1 (data not shown). Taken together these data suggest that the ‘Tyr-finger’ in importin α5, at position 476 is critical for unfolding of ARM 10. As seen in the structure of importin α5 bound to influenza virus polymerase PB2 32, ARM 10 helix H3 ‘embraces’ the import cargos and retains it bound to importin α5. Although Try476 is necessary to confer pSTAT1 binding to importin α5, this residue is not sufficient to turn importin α1 into a pSTAT1 binder. Isoform-specific differences in addition to ARM 10 flexibility are likely responsible for high affinity binding to pSTAT1.
Double stranded DNA displaces importin α5 from pSTAT1
In a last set of experiments, we asked whether double stranded DNA (dsDNA) could directly displace importin α5 from pSTAT1, thereby dissociating the pSTAT1 import complex. It was previously reported that binding of activated STAT1 to GST-importin α5 could be displaced through the addition of an excess of dsDNA corresponding to the IFN-γ activated DNA site (GAS) 37; 38. However, these analyses were done with whole-cell lysates expressing pSTAT1, as opposed to homogeneously purified pSTAT1, and GST-fused importin α5 immobilized over glutathione beads. In our experimental set up, we aimed at measuring the direct displacement of importin α5 by DNA in solution, using purified factors. To test this idea, we developed a simple displacement assay on native agarose gel. Both importin α5 and pSTAT1 are readily visible on agarose gel as single bands (Fig. 5A, lane 1 and 2). Likewise, a gel-filtration purified pSTAT1:importin α5 complex appears greatly shifted toward the negative pole as compared to free pSTAT1 (Fig. 5A, lane 3). To check for importin α5 displacement by dsDNA, 60 μg of a preformed (gel-filtration purified) pSTAT1:importin α5 complex (Fig 5A, lane 3) was incubated in solution with increasing concentrations (between 0.5 to 2-fold molar excess) of a 38-mer dsDNA oligonucleotide containing two tandem cfosM67-promoter elements. This oligonucleotide is known to bind pSTAT1 with high affinity 2 (Fig. 5A, lane 7), and results in a pSTAT1:DNA complex migrating on agarose gel slightly below the pSTAT1:importin α5 complex (compare Fig. 5A lane 7 and 3, respectively). After incubation at room temperature, mixtures containing pSTAT1, importin α5 and DNA were separated on agarose gel (Fig 5A, lanes 4-6). In agreement with the previous data, we found that the addition of dsDNA resulted in the displacement of increasing quantities of importin α5 from pSTAT1. The former is visible on agarose gel as a thin band migrating below the pSTAT1:DNA complex (Fig 5A, lanes 4-6). To determine the efficiency of dsDNA-mediated displacement of importin α5, we first estimated the total quantity of importin α5 bound to pSTAT1 in the starting sample. Due to the size of importin α5 (M.W. ~65 versus M.W. 165 kDa of pSTAT1) and the 2:1 stoichiometry of the pSTAT1:importin α5 complex, importin α5 represents only 27.5% of the molecular mass of the pSTAT1:importin α5 complex (visible in Fig. 5A, lane 3 without dsDNA), corresponding to ~16.5 μg of the total 60 μg loaded on gel. To determine the amount of importin α5 displaced by dsDNA in Fig 5A, lane 4-6, we generated a standard curve using known amounts of importin α5, which were separated under identical conditions, on the same native agarose gel (Fig. 5B). Densitometric quantification of native agarose gel bands showed a linear response to the amount of importin α5 loaded on gel (Fig. 5C). To confirm the accuracy of this calibration curve, the control importin α5 in Fig. 5A, lane 1 (corresponding to 24 μg of protein) was also quantified densitometrically and, using the calibration curve in Fig. 5C, estimated to contain ~22 μg of importin α5, very close to the quantity loaded on gel. Likewise, the amount of importin α5 displaced by dsDNA was also quantified using the calibration curve. We found that dsDNA displaced approximately 8 μg, 13.5 μg, and 14.2 μg of importin α5 when added at molar ratios (DNA:importin α5) of 0.5:1, 1:1, and 2:1 (Fig. 5A, lanes 4-6). Thus, equimolar concentration of dsDNA can displace nearly 82% (or 13.5 μg) of the 16.5 μg of importin α5 bound to pSTAT1, strengthening the idea that importin α5 and dsDNA occupy mutually exclusive binding sites on pSTAT1.
Figure 5. A DNA oligonucleotide containing two cfosM67-promoter elements can displace importin α5 from pSTAT1.
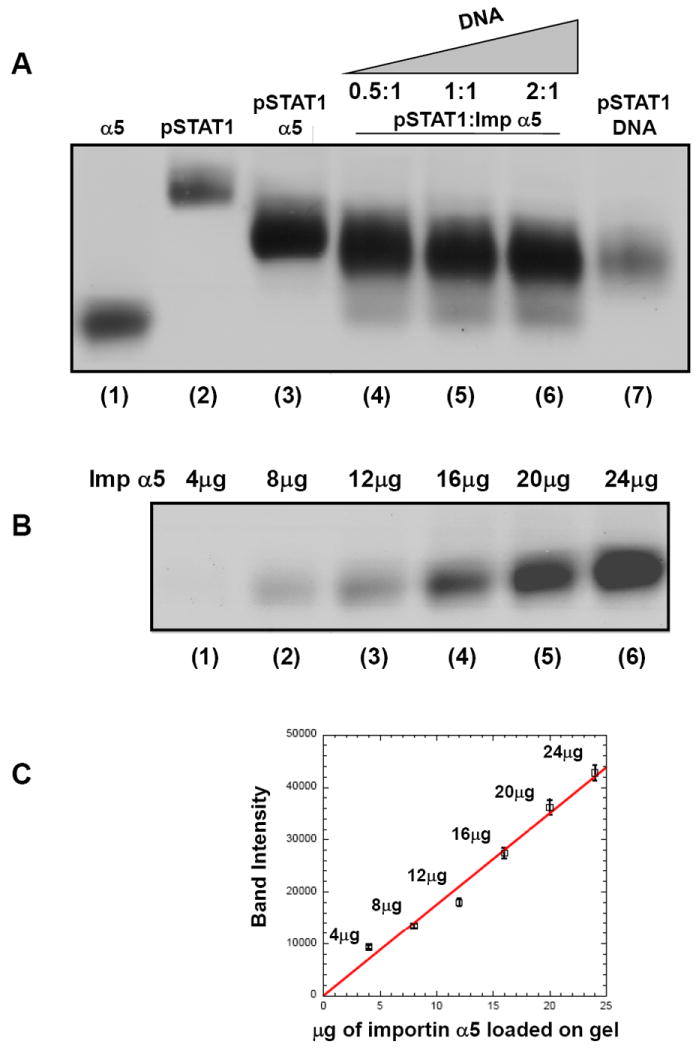
Electrophoretic mobility shift assay on native agarose gel. Free importin α5 (24 μg), pSTAT1 (25 μg) and the pSTAT1:α5 complex (60 μg) are in lanes 1, 2 and 3, respectively. Addition of 0.5-, 1.0-, 2.0-fold molar excess (over importin α5) of a 38-mer dsDNA oligonucleotide containing two cfosM67-promoter elements to 60 μg of pSTAT1:α5 displaces importin α5 from pSTAT1 and yields a new pSTAT1:DNA complex (in lanes 4, 5, 6, respectively). A control of the pSTAT1:DNA complex formed by adding an excess of DNA to 25 μg pSTAT1 is in lane 7. (B) Importin α5 titration on native agarose gel. 4, 8, 12, 16, 20, and 24 μg of importin α5 were loaded in lanes 1-6, respectively. (C) Standard curve calculated for the intensity of importin α5 bands in panel B. The band intensity was quantified densitometrically. A line of best fit was calculated using the ordinary least squares method in KaleidaGraph ®. The error bars represent the standard deviation of three independent quantifications.
DISCUSSION
STATs form an important class transcription factors that mediate transduction of extracellular stimuli from the cell surface to the cell nucleus 1. Efficient and rapid translocation of STAT1 in response to γ-IFN stimulation is essential to ensure expression of cytokine-dependent genes. In this study, we have expanded our understanding of the molecular basis for recognition of pSTAT1 by the transport adaptor importin α5. Using sedimentation velocity analysis we demonstrated that a single equivalent of importin α5 molecule binds a dimer of phosphorylated STAT1, contradicting previous reports (using Western blot analysis) that GST-fused importin α5 interacts with activated STAT1 in a 1:1 molar ratio 38. Additionally, using ITC we demonstrated that phospho-Tyr701, in activated STAT1, although essential for binding to importin α5, is not a direct binding determinant. This observation strongly suggests that recognition of pSTAT1 by importin α5 is distinct from that of EBNA-1, which is preferentially bound and imported upon Ser-phosphorylation at position 385 41. Furthermore, we provide evidences that importin α5 ARM 10 and not the C-terminal tail (res. 512-538), contains a critical determinant for pSTAT1 binding. This determinant can be transferred in trans to importin α1, which has negligible binding affinity for pSTAT1. By comparing the primary sequences and tertiary structures of importin α5 and α1 we identified a single Tyr residue at position 476 that is conserved in the three members of the NPI-1-family, (α5, α6 and α7). Based on the structure of importin α5 32, we hypothesized Tyr476 inserts in between ARM 10 helices H1, H2, H3 like a ‘Tyr-finger’ to destabilize the inter-helical folding of ARM 10. This, in turn, results in a mobile helix H3, which, in members of the NPI-1 family, may be used to swing around the import substrate (as shown for importin α5 binding to influenza subunit PB2) 32. To support this hypothesis, mutation of Tyr476 to Gly completely abolished binding of importin α5 to pSTAT1, thereby lending support to the idea that importin α5 recognizes PB2 and pSTAT1 with fundamentally similar mechanisms involving ARM 10. However, the reverse mutation in importin α1, Gly468Tyr, did not result in a gain of function in pSTAT1 binding, suggesting that the flexibility of ARM 10 H3 helix is necessary but not sufficient for high affinity binding to pSTAT1.
A structural model of importin α5 bound to pSTAT1
The analogy between influenza RNA polymerase PB2 and pSTAT1 recognition by importin α5 prompted us to investigate whether there are structural similarities between STAT1 and the C-terminal domain of influenza subunit PB2. Unexpectedly, superimposition of influenza subunit PB2 (pdb 2JDQ) to pSTAT1 core (pdb 1BF5) revealed striking structural similarity between the PB2 globular domain (res. 686-757) and pSTAT1 SH2-domain (res. 558-634) (Fig. 6A). The root mean square deviation (rmsd) deviation (on the α-carbon) between these two domains is ~2.3Å, and although the SH2-domain is significantly larger, the PB2 structural core formed by three β-strands and an α-helix (β1−α2−β3−β4) superimposes well onto the STAT1 SH2-domain (Fig. 6A).
Figure 6. A structural model for the pSTAT1:importin α5 nuclear import complex.
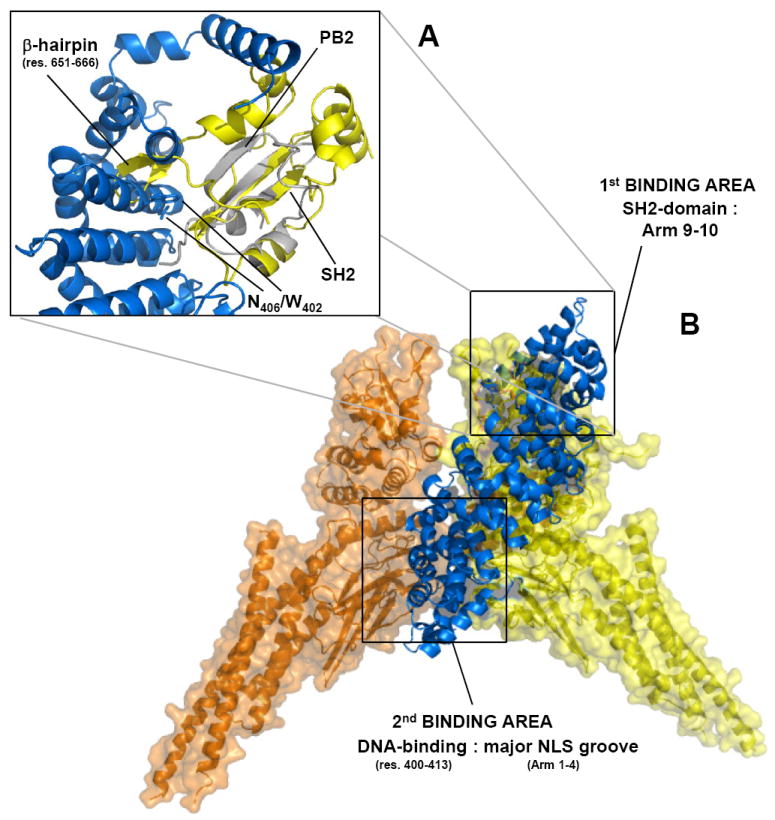
(A) Superimposition of influenza PB2 subunit bound to importin α5 (in gray and blue, respectively) with STAT1–SH2 domain (in yellow). The rmsd deviation (on the α-carbon) between the PB2 globular domain (res. 686-757) and pSTAT1 SH2-domain (res. 558-634) is ~2.3 Å. (B) Structural model of dimeric pSTAT1 bound to one equivalent of importin α5. Importin α5 is shown as ribbon and colored in blue. Dimeric pSTAT1 is also in depicted in ribbon that is overlaid to a semi-transparent surface colored in orange and yellow for the two identical pSTAT1 protomers.
Although the crystal structure of pSTAT1 in complex with importin α5 is not currently available, the similarity between influenza PB2 subunit and STAT1-SH2 domain and the involvement of ARM 10 local unfolding demonstrated in this work provides a structural framework to model the interaction of pSTAT1 with importin α5. In addition, a large body of mutational data is available in the literature, which can be used as structural ‘restraints’ to improve the accuracy of a structural model between pSTAT1 and importin α5. In particular, we focused on the following published data as well as findings reported in this paper: (i) STAT1 binds importin α5 only as a phosphorylated dimer 34; (ii) binding of pSTAT1 to importin α5 requires STAT1 N-terminal domain (res. 1-132) 39; (iii) pSTAT1 N-terminal domains do not need to interact for importin α5-mediated STAT1 nuclear import 48; (iv) region 400-413 of STAT1, located in the DNA binding domain and known as dimer specific NLS (dsNLS) is essential for importin α5 binding 35; 37; 38; (v) acid substitutions in ARM repeat 8 (Trp402Ala and Asp406Ala) or 9 (Asp445Ala and Asp449Ala) prevent pSTAT1 binding to importin α5 46; (vi) and the stoichiometry of the pSTAT1:α5 import complex is 2:1 (shown here).
Fig. 6B presents a structural model of the pSTAT1:importin α5 complex that satisfies all biochemical and structural data presented above. Importin α5 is proposed to bind asymmetrically in between two STAT1 monomers. Dimeric pSTAT1 engages in importin α5 binding in a parallel conformation, with the transport adaptor deeply inserted in the groove between two STAT1 protomers, in a position otherwise occupied by DNA in the nuclear conformation 7. Two major surface binding areas in pSTAT1, located within the SH2- and DNA-binding domains of opposite STAT1 protomers, contact the importin α5 C-terminal ARM 9-10 and the major NLS binding groove (ARM 1-4), respectively (Fig. 6B). In the first binding area (Fig. 6A), pSTAT1-SH2 is embraced by ARM 10 helix H3 and is located in our model in close proximity to ARM 8-9 residues, essential for pSTAT146. A β-hairpin in STAT1 SH2-domain spanning residues 651-666 must move by several Angstroms in response to importin α5 binding from the initial position occupied in the structure of pSTAT1 core bound to DNA 7 (Fig. 6A). This movement would position the STAT1-β-hairpin in close proximity to importin α5 ARM 9 residues Trp402 and Asn406, which are known to be important for pSTAT1 binding 46 (Fig. 6A). In the second binding area (Fig. 6B), the importin α5 major NLS binding groove (ARM 1-4) is positioned within bonding distance from the DNA binding domain of an opposite STAT1 protomer. While, in the absence of a crystal structure, it is impossible to predict with absolute certainty the exact residues engaging in binding interactions, it is possible that the entire importin α5 surface between ARM 2-6, where arrays of Trp/Asn are conserved 46, may be involved in contacting basic pSTAT1 side chains.
The structural model proposed in Fig. 6B is in a good agreement with four biochemical observations made in this paper. First, asymmetry in STAT1:importin α5 recognition explains the 2:1 stoichiometry observed by sedimentation velocity analysis (Fig. 1C). Since pSTAT1 has a twofold symmetric axis running along the two protomers, the asymmetric binding of importin α5 ensures binding to a dsNLS in the DNA-binding domain of one pSTAT1 protomer, while burying the other STAT1 protomer DNA-binding domain. This prevents high affinity binding of a second monomer of importin α5 to pSTAT1. Second, the semi-buried position of importin α5 inside dimeric pSTAT1 is not expected to increase STAT1 hydrodynamic radius, which is consistent both with the nearly identical frictional ratio of pSTAT1 dimer and the pSTAT1:importin complex (Fig. 1C, right panel), and their similar migration during gel-filtration chromatography (Fig 1A). Third, ARM 10 helix H3 in importin α5 is proposed to bind pSTAT1-SH2 domain, thereby locking the import substrate into a conformation competent for binding to pSTAT1 dsNLS located in the DNA binding domain. The function of this ‘molecular-embracing’ is likely to prevent cargo dissociation. To support this idea, the point mutation Tyr476Gly in importin α5 dramatically reduced the affinity for pSTAT1 to the micromolar range mainly by increasing the importin α5 off-rate from pSTAT1 (koff = 0.289s-1 versus 0.0183s-1 in wild type importin α5 Table I). Fourth, the position of importin α5 inside the pSTAT1 DNA binding grove is in excellent agreement with the observation that dsDNA can displace importin α5 from pSTAT1 in vitro (Fig. 5A). Possibly other factors contribute in vivo to disassembly of the STAT1 import complex and release of activated STAT1 in the cell nucleus. For instance, Nup50 was shown to bind the C-terminus of importin α1 to increase the rate of NLS-cargo release from the adaptor 49. Although direct interaction of Nup50 with importin α5 has not been demonstrated, Nup50 binds the C-terminal region of importin α1 with high affinity, in a region that overlaps the mapped STAT1 binding site in importin α5. Thus, Nup50 seems a primary candidate for STAT1 import complex disassembly. Likewise, the export factor CAS may compete with importin β for binding to importin α5, thereby providing an additional way to disassemble the pSTAT1-import complex and release pSTAT1 in the cell nucleus.
In conclusion, the recognition of pSTAT1 by importin α5 appears to be more complex than the well documented molecular binding of basic NLS peptides by importin α1 22; 24; 25; 50. The binding constant of importin α5 for pSTAT1 is Kd ~191 ± 20 nM, which is ~10-fold lower compared with most classical NLS-cargos binding to importin α1 (generally in a range of Kd ~ 1-20 nM) 51. However, in the absence of an autoinhibitory mechanism by the importin α5 IBB-domain (as demonstrated in this paper), this affinity is sufficiently high to allow for efficient nuclear import of pSTAT1. Furthermore, unlike classical NLSs, the recognition of pSTAT1-NLS by importin α5 is less likely competed off by non-specific interactions with ‘wrong’, broadly basic sequences that resemble NLSs 52. Thus, despite the lower Kd, the recognition of STAT1 by importin α5 is both specific and kinetically effective. Interestingly, the asymmetric recognition of phosphorylated STAT1 by importin α5 described in this paper presents significant analogies to the recognition of the transcription factor NF-κB p50:p65 by importin α3. Fagerlund et al. observed that a heterodimer of p50:p65 is simultaneously bound by different NLS binding sites of importin α3 53. Whereas p50 interacts with the adaptor N-terminus, p65 binds the importin α3 C-terminal NLS binding site. Similarly to pSTAT1, the stoichiometry of the NF-κB:importin α3 import complex is consistent with 2:1. Unlike STAT1, however, p50 and p65 are related, yet not identical proteins. This suggests that isoforms importin α3 and α5 can recognize asymmetric epitopes in dimeric transcription factors, which allows these karyopherins to stabilize complex import cargos during nuclear import. In conclusion, further structural and biochemical work will be necessary to fully understand the molecular basis for the asymmetric recognition and nuclear import of non-classical dimeric import substrates like STAT1 and NF-KB.
MATERIAL AND METHODS
Synthetic peptides and oligonucleotide
Peptides corresponding to EBV EBNA-1 NLS (379-KRPRSPpSS-386), EBNA-1 phospho-NLS (379-KRPRSPSS-386), and a phospho-STAT1 peptide spanning residues 695-708 (695-G PKGTGpYIKTELIS-708) were synthesized and purified by Genscript. A 38-mer double stranded DNA oligonucleotide containing two tandem cfosM67-promoter elements spaced by 10 nucleotides (5’-ACGGTTTCCCGTAAATTGACGGATTTCCCGTAAATGGC-3’) was purchased from Biotez GmbH, Berlin.
Plasmids and DNA constructs
The cDNA encoding full length STAT1 or STAT1 lacking residues 1-132 (STAT1 core) was cloned between the Kpn1 and EcoR1 restriction sites of an engineered PMAL-c2e vector lacking the MBP gene and containing an N-terminal 6-histidine tag. Importin α5 and α1 were cloned in pGEX-6p-2 vectors. All related constructs importin ΔIBB-α5 (res. 79-538), α5 (res. 1-512), α5 (res. 1-504), α5 (Y476G) and α1 (G468Y) were generated using QuickChange™ site-directed mutagenesis. The importin α5/α1 hybrid consisted of nucleotides 1-1347 of importin α5 followed by nucleotides 1339-1587 of importin α2, inserted between the BamH1 and Not1 sites of pGEX-6p-2 vector. The α1/α5 hybrid consisted of nucleotides 1-1338 of importin α1 followed by nucleotides 1348-1614 of importin α5, inserted between the Sal1 and Not1 sites of pGEX-4T-3 vector. Strep-tagged STAT1 insect cell expression clone was described 4. Full-length importin-α5 with C-terminal Strep-tag was expressed from pASK-IBA3. All constructs generated in this study were fully sequenced to ensure the correctness of the DNA sequence.
Protein expression and purification
Expression of full length STAT1 and STAT1 core in E. coli strain BL21-CodonPlus (DE3) RIL (Stratagene, La Jolla, CA) was performed under the control of an IPTG-inducible promoter for 5 hrs at 30° C. Both STAT1 constructs were N-terminally fused to a 6-histidine tag, and purified by metal chelate affinity chromatography. Alternatively, STAT1 C-terminally fused to Strep-tag was expressed in insect cells as previously described 4. STAT1 was alkylated with N-ethyl-maleimide and in-vitro phosphorylated with purified EGF receptor as previously described 2. Unphosphorylated STAT1 was separated from phosphorylated STAT1 by heparin chromatography. pSTAT1 constructs were further purified by size exclusion chromatography over a Superdex 200 column (GE Healthcare) equilibrated in buffer G.F. (20 mM Tris pH 8.0, 150 mM NaCl, 1 mM TCEP, 1 mM EDTA, and 0.2 mM PMSF). Strep-tagged importin α5 was expressed in E.coli strain BL21-CodonPlus (DE3) RIL for 3 hrs at 30° C and purified using Strep tactin-sepharose according to the manufacturer’s recommendations (IBA GmbH, Göttingen). Recombinant GST-fused importin α5 was expressed in E.coli strain BL21-CodonPlus (DE3) RIL and purified as previously described 34. The GST-tag was removed using Prescission Protease, and separated by anion exchange chromatography using a stepwise salt gradient over DEAE resin. Importin α5 was further purified by size exclusion chromatography over a Superdex 200 column (GE Healthcare) equilibrated in buffer G.F.
Biochemical techniques
Analytical gel-filtration was done with Superdex 200 10/300 GL in PBS. Pull down assay were carried out as previously described 54. For the pull down assay, purified recombinant GST-tagged importin α5 (res. 1-538) and the importin α5 Tyr476Gly mutant were immobilized separately on Glutathione Sepharose 4B resin (GE Healthcare). 250 μg of pSTAT1 was incubated with the GST-importin αs for 1hr on ice, and washed thoroughly. SDS-PAGE and Western Blot analysis were performed, utilizing an HRP-conjugated penta-His antibody (Qiagen) to detect pSTAT1, strep-tagged proteins were detected with anti-strep-antibody (IBA GmbH Göttingen). The native band-shift assay on agarose gel was carried out as previously described 54. Importin α5 (24 μg), pSTAT1 (25 μg), and a gel-filtration purified complex of importin α5 bound to pSTAT1 (60 μg) were separated in a 2% agarose gel at 4° C for 5 hrs for the native band shift assay. To test DNA-dependent displacement of importin α5, 60 μg of gel-filtration purified pSTAT1:importin α5 complex was incubated with 0.5-, 1.0- and 2.0-fold molar excess (over importin α5) of the 38-mer dsDNA oligonucleotide cfosM67-promoter element, for 30 minutes on ice prior to electrophoretic separation on agarose gel. After electrophoresis, the native gel were fixed in Gel Fixing solution (25% (v/v) isopropanol and 10% (v/v) acetic acid) for 20 minutes and then equilibrated with 95% (v/v) ethanol for 2 hrs. Gels were then dried on a piece of filter paper, stained for 10 minutes in 0.4% (w/v) Coomassie brilliant blue R250 in Gel Fixing solvent and destained in Gel Fixing solvent until the background was clear. Standard curve to quantify importin α5 was generated by running 4, 8, 12, 16, 20, and 24 μg of importin α5 in the same gel used for DNA displacement assay. NIH Image J software 55 was used to analyze band intensities, which were plotted against the known concentration of protein loaded in each lane. A line of best fit was calculated using the ordinary least squares method in KaleidaGraph ®. The error bars represent the standard deviation of three independent bands of importin α5 containing the same quantity of protein and analyzed on agarose gel under identical conditions.
Analytical ultracentrifugation
All experiments were performed using a Beckman Optima XL-A at the Kimmel Cancer Center (KCC) X-ray Crystallography and Molecular Characterization shared resource facility. All samples were analyzed in absorbance mode at 280 nm in buffer GF, containing 20 mM Tris, pH 8, 150 mM NaCl, 1 mM EDTA, and 1 mM TCEP. The data were collected at 10 °C, and at 30.000 rpm, using standard epon 2-channel centerpieces and an An-50 Ti rotor. The following loading concentrations were used: importin α5, 1 mg/ml; pSTAT1, 0.7 mg/ml; uSTAT1, 0.8 mg/ml; and a gel-filtration purified complex of importin α5:pSTAT1, 0.6 mg/ml. Hydrodynamic corrections for buffer density and viscosity were made as implemented in UltraScan 56. Data were analyzed with the UltraScan software suite, v9.9 57. Molecular weight and shape distributions obtained in the two-dimensional spectrum analysis 58 were further refined using genetic algorithm 59. Statistics were subjected to 50 cycles of Monte Carlo analysis 60. All calculations were performed on the Laredo cluster at the Bioinformatics Core Facility at the University of Texas Health Science Center at San Antonio.
Isothermal titration calorimetry (ITC)
ITC experiments were carried out at 30° C in a VP-ITC calorimeter (Microcal). A phosphorylated EBNA-1 peptide dissolved in ITC buffer (10 mM HEPES pH 7.4, 150 mM sodium chloride, 1 mM EDTA, 1 mM TCEP) at a concentration of 450 μM was injected in 10 μL increments into the calorimetric cell containing 1.8 ml of importin α5 at a concentration of 50 μM. The spacing between injections was 300 seconds. In separate experiments, the unphosphorylated EBNA-1 peptide and phospho-Tyr STAT1 peptide were titrated into importin α5 as described above. Titration data were analyzed using the Origin 7.0 data analysis software (Microcal Software, Northampton, MA). Injections were integrated following manual adjustment of the baselines. Heats of dilution were determined from control experiments with the ITC buffer and subtracted prior to curve fitting using a single set of binding sites model. The curve fitting yields a Kd= 3 ± 14 μM and ΔH = 6948 ± 1491 cal/mol at 30°C for titration of the phosphorylated EBNA-1 peptide into importin α5 (79-538), and a Kd ~30 ± 15 μM and ΔH = 6548 ± 1191 cal/mol for titration of the unphosphorylated EBNA-1 peptide into importin α5.
Surface plasmon resonance (SPR)
SPR experiments were carried out on a Biacore 3000 instrument, equilibrated at 25°C in HBS-EP buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% Surfactant P20; GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Purified recombinant GST-α5 was captured on a CM5 sensor chip coupled with immobilized goat anti-GST antibody. Samples of pSTAT1 were applied to the chip at various concentrations between 30-500 nM in HBS-EP buffer. Response units were recorded at a flow rate of 30 μl min-1 with injection time and wait time after injection of 3 and 10 min, respectively. Each experiment was repeated 4 times. Experiments were performed as stated above with GST-tagged full length importin α5, ΔIBB-α5 (res. 79-538), α5(res. 1-512), α5(res. 1-504), α1/α5 hybrid, and α5/α1 hybrid constructs. Recombinant pSTAT1 core and unphosphorylated STAT1 were applied to chip-immobilized GST-full length importin α5 at concentrations between 30-1000 nM. The apparent association and dissociation rate constants and χ2 values were calculated using the conformational change model in BIAevaluation software Version 4.1 (Biacore AB). A complete list of kinetic parameters is in Table I.
Molecular modeling
Superimposition of importin α5 bound to the C-terminal domain of influenza virus polymerase PB2 (pdb #2JDQ) with STAT1 (pdb #1BF5 and #1YVL) was achieved using the CCP4 61 program Superimpose. A 3D-dimensional model of pSTAT1 bound to importin α5 was built in Coot 62.
Acknowledgments
We thank Kaylen Lott and Anshul Bhardwaj for technical help, stimulating discussions and critical reading of the manuscript; and Edda Schulz for providing importin α5 expression construct. This work was supported by NIH grant GM074846-01A1 to G.C; and DFG grant VI 218/2 and BBSRC grant BB/G019290/1 to U.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 2.Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE., Jr DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. Embo J. 1996;15:5616–26. [PMC free article] [PubMed] [Google Scholar]
- 3.Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–7. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 4.Wenta N, Strauss H, Meyer S, Vinkemeier U. Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc Natl Acad Sci U S A. 2008;105:9238–43. doi: 10.1073/pnas.0802130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, McMurray JS, Demeler B, Darnell JE, Jr, Chen X. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–71. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 6.Ota N, Brett TJ, Murphy TL, Fremont DH, Murphy KM. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat Immunol. 2004;5:208–15. doi: 10.1038/ni1032. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–39. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 8.ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, Zhu W, Tremblay M, David M, Shuai K. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22:5662–8. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer T, Marg A, Lemke P, Wiesner B, Vinkemeier U. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev. 2003;17:1992–2005. doi: 10.1101/gad.268003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan KL, Broyles SS, Dixon JE. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature. 1991;350:359–62. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- 11.Najarro P, Traktman P, Lewis JA. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses Stat1 activation. J Virol. 2001;75:3185–96. doi: 10.1128/JVI.75.7.3185-3196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koksal AC, Nardozzi JD, Cingolani G. Dimeric quaternary structure of the prototypical dual specificity phosphatase VH1. J Biol Chem. 2009;284:10129–37. doi: 10.1074/jbc.M808362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 14.Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–56. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–98. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 16.Weis K. Nucleocytoplasmic transport: cargo trafficking across the border. Curr Opin Cell Biol. 2002;14:328–35. doi: 10.1016/s0955-0674(02)00337-x. [DOI] [PubMed] [Google Scholar]
- 17.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–88. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lott K, Bhardwaj A, Mitrousis G, Pante N, Cingolani G. The importin beta binding domain modulates the avidity of importin beta for the nuclear pore complex. J Biol Chem. 2010;285:13769–80. doi: 10.1074/jbc.M109.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–14. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 20.Andrade MA, Petosa C, O’Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 21.Kohler M, Speck C, Christiansen M, Bischoff FR, Prehn S, Haller H, Gorlich D, Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol Cell Biol. 1999;19:7782–91. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 23.Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–38. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 24.Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol. 2000;297:1183–94. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 25.Fontes MR, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J Biol Chem. 2003;278:27981–7. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- 26.Chen MH, Ben-Efraim I, Mitrousis G, Walker-Kopp N, Sims PJ, Cingolani G. Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin alpha. J Biol Chem. 2005;280:10599–606. doi: 10.1074/jbc.M413194200. [DOI] [PubMed] [Google Scholar]
- 27.Yang SN, Takeda AA, Fontes MR, Harris JM, Jans DA, Kobe B. Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-alpha using oriented peptide library screening. J Biol Chem. doi: 10.1074/jbc.M109.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol. 1999;6:388–97. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 29.Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J Biol Chem. 2001;276:1317–25. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 30.O’Neill RE, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–25. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 31.Meyer T, Vinkemeier U. Nucleocytoplasmic shuttling of STAT transcription factors. Eur J Biochem. 2004;271:4606–12. doi: 10.1111/j.1432-1033.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 32.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol. 2007;14:229–33. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 33.Gorlich D, Seewald MJ, Ribbeck K. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. Embo J. 2003;22:1088–100. doi: 10.1093/emboj/cdg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. Embo J. 1997;16:7067–77. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer T, Begitt A, Lodige I, van Rossum M, Vinkemeier U. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 2002;21:344–54. doi: 10.1093/emboj/21.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J Biol Chem. 2001;276:16447–55. doi: 10.1074/jbc.M008821200. [DOI] [PubMed] [Google Scholar]
- 37.McBride KM, Banninger G, McDonald C, Reich NC. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. Embo J. 2002;21:1754–63. doi: 10.1093/emboj/21.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fagerlund R, Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J Biol Chem. 2002;277:30072–8. doi: 10.1074/jbc.M202943200. [DOI] [PubMed] [Google Scholar]
- 39.Meissner T, Krause E, Lodige I, Vinkemeier U. Arginine methylation of STAT1: a reassessment. Cell. 2004;119:587–9. doi: 10.1016/j.cell.2004.11.024. discussion 589-590. [DOI] [PubMed] [Google Scholar]
- 40.Marg A, Shan Y, Meyer T, Meissner T, Brandenburg M, Vinkemeier U. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J Cell Biol. 2004;165:823–33. doi: 10.1083/jcb.200403057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitamura R, Sekimoto T, Ito S, Harada S, Yamagata H, Masai H, Yoneda Y, Yanagi K. Nuclear import of Epstein-Barr virus nuclear antigen 1 mediated by NPI-1 (Importin alpha5) is up- and down-regulated by phosphorylation of the nuclear localization signal for which Lys379 and Arg380 are essential. J Virol. 2006;80:1979–91. doi: 10.1128/JVI.80.4.1979-1991.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Bhandari R, Vinkemeier U, Van Den Akker F, Darnell JE, Jr, Kuriyan J. A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci. 2003;12:361–5. doi: 10.1110/ps.0218903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harreman MT, Cohen PE, Hodel MR, Truscott GJ, Corbett AH, Hodel AE. Characterization of the auto-inhibitory sequence within the N-terminal domain of importin alpha. J Biol Chem. 2003;278:21361–9. doi: 10.1074/jbc.M301114200. [DOI] [PubMed] [Google Scholar]
- 44.Harreman MT, Hodel MR, Fanara P, Hodel AE, Corbett AH. The auto-inhibitory function of importin alpha is essential in vivo. J Biol Chem. 2003;278:5854–63. doi: 10.1074/jbc.M210951200. [DOI] [PubMed] [Google Scholar]
- 45.Herold A, Truant R, Wiegand H, Cullen BR. Determination of the functional domain organization of the importin alpha nuclear import factor. J Cell Biol. 1998;143:309–18. doi: 10.1083/jcb.143.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melen K, Fagerlund R, Franke J, Kohler M, Kinnunen L, Julkunen I. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J Biol Chem. 2003;278:28193–200. doi: 10.1074/jbc.M303571200. [DOI] [PubMed] [Google Scholar]
- 47.Ma J, Cao X. Regulation of Stat3 nuclear import by importin alpha5 and importin alpha7 via two different functional sequence elements. Cell Signal. 2006;18:1117–26. doi: 10.1016/j.cellsig.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 48.Meyer T, Hendry L, Begitt A, John S, Vinkemeier U. A single residue modulates tyrosine dephosphorylation, oligomerization, and nuclear accumulation of stat transcription factors. J Biol Chem. 2004;279:18998–9007. doi: 10.1074/jbc.M400766200. [DOI] [PubMed] [Google Scholar]
- 49.Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24:3681–9. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure Fold Des. 2000;8:329–38. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 51.Catimel B, Teh T, Fontes MR, Jennings IG, Jans DA, Howlett GJ, Nice EC, Kobe B. Biophysical characterization of interactions involving importin-alpha during nuclear import. J Biol Chem. 2001;276:34189–98. doi: 10.1074/jbc.M103531200. [DOI] [PubMed] [Google Scholar]
- 52.Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang W, Chait BT, Rout MP. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J Cell Biol. 2006;175:579–93. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fagerlund R, Kinnunen L, Kohler M, Julkunen I, Melen K. NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem. 2005;280:15942–51. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- 54.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J Biol Chem. 2008;283:7877–84. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 55.Rasband WS. U. S. National Institutes of Health; Bethesda, Maryland, USA: 1997-2009. http://rsb.info.nih.gov/ij/ [Google Scholar]
- 56.Demeler B. Analytical Ultracentrifugation as a Key Complementary Technique in Structural Biology. In: Scott D, Harding SE, Rowe A, editors. Modern Analytical Ultracentrifugation: Techniques and Methods. Royal Society of Chemistry; Cambridge, UK: 2005. pp. 210–229. [Google Scholar]
- 57.Demeler B. UltraScan: a comprehensive data analysis software package for analytical ultracentrifugation experiments. 2010 http://www.ultrascan.uthscsa.edu.
- 58.Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39:405–14. doi: 10.1007/s00249-009-0413-5. [DOI] [PubMed] [Google Scholar]
- 59.Brookes E, Demeler B. GECCO Proceedings. ACM; New York: 2007. Parsimonious regularization using genetic algorithms applied to the analysis of analytical ultracentrifugation experiments. [Google Scholar]
- 60.Demeler B, Brookes E. Monte Carlo analysis of sedimentation experiments. Colloid Polym Sci. 2008;286:129–137. [Google Scholar]
- 61.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]


