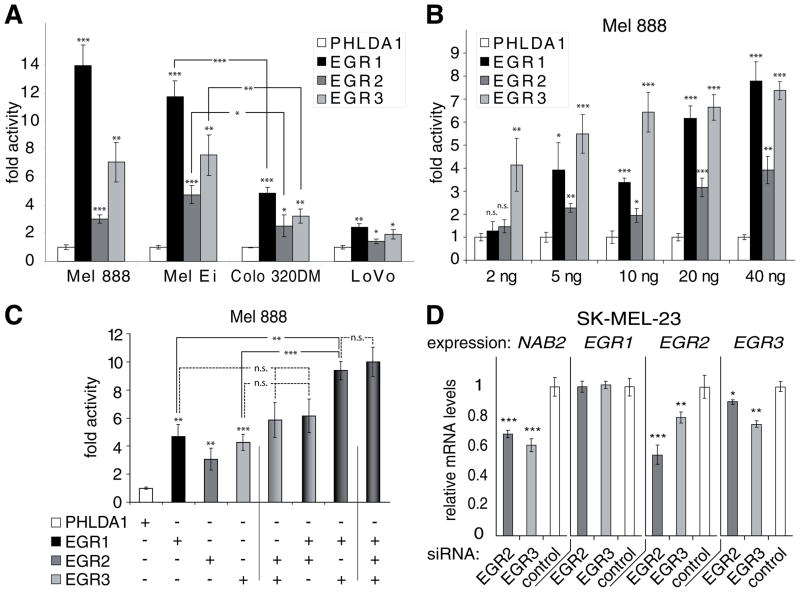
Fig. 3.
EGR1, EGR2, and EGR3 are important regulators of NAB2 transcription. A. EGR1, EGR2, and EGR3 activate the NAB2 promoter. Mel 888, Mel Ei, Colo 320DM, and Lovo cells were transfected with 60 ng of maluc-679, which contains the human NAB2 promoter, and 40 ng of either human EGR1, EGR2, EGR3, or PHLDA1 control DNA. Luciferase activity was determined as described in “Experimental Procedures”. Results are presented as fold activity over luciferase activity of maluc-679 cotransfected with PHLDA1. Average and standard deviation of triplicates from a representative experiment is shown. * P <0.05, ** P <0.01, and *** P <0.001 of PHLDA1 versus EGR1, EGR2, EGR3 or as indicated by connecting lines. P values were calculated using Student’s t-test. B. EGR1, EGR2, and EGR3 activate the NAB2 promoter in a dose-dependent manner. Mel 888 cells were transfected with 60 ng of maluc-679, which contains the human NAB2 promoter, and increasing amounts, as indicated, of either human EGR1, EGR2, EGR3, or PHLDA1 control DNA. Empty vector DNA added to make a total of 100 ng. Luciferase activity was determined as described in “Experimental Procedures”. Results are presented as fold activity over luciferase activity of maluc-679 cotransfected with PHLDA1. Average and standard deviation of triplicates from a representative experiment is shown. * P <0.05, ** P <0.01, *** P <0.001, n.s., not significant (PHLDA1 versus EGR1, EGR2, or EGR3). P values were calculated using Student’s t-test. C. EGR1 and EGR3 act in concert to activate the NAB2 promoter. Mel 888 cells were transfected with 60 ng of maluc-679 and different combinations, as indicated, of 15 ng of human EGR1, EGR2, EGR3, or PHLDA1 control plasmid DNA. Vector DNA was added to make a total of 105 ng. Luciferase activity was determined as described in “Experimental Procedures”. Results are presented as fold activity over luciferase activity of maluc-679 cotransfected with PHLDA1 control plasmid DNA. Average and standard deviation of triplicates from a representative experiment is shown. ** P <0.01, *** P <0.001, n.s., not significant of PHLDA1 versus EGR1, EGR2, EGR3 or as indicated by connecting lines. P values were calculated using Student’s t-test. D. Depletion of EGR2 or EGR3 reduces NAB2 expression in the human melanoma cell line SK-MEL-23. SK-MEL-23 cells were transfected with negative control siRNA or siRNAs directed against EGR2 or EGR3 as described in “Experimental Procedures”. 24 hours later mRNA expression of NAB2, EGR1, EGR2, and EGR3 was determined using real time PCR (as described in “Experimental Procedures”). Average mRNA expression in relation to negative control (black bar) and standard deviation of triplicates from one representative experiment out of two performed is shown. * P <0.05, ** P <0.01, and *** P <0.001 of control siRNA versus siRNA against EGR2 or EGR3. P values were calculated using Student’s t-test.