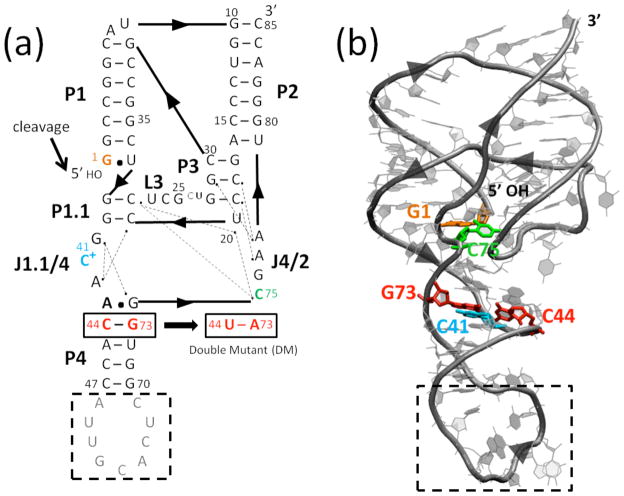
Fig. 1.
Structure of the genomic HDV ribozyme. (a) Secondary and (b) tertiary structures of the cleaved (product) form of the genomic HDV ribozyme. Arrowheads denote 5′ to 3′ directionality. (a) Ribozyme is comprised of five pairing regions, P1–P4 and P1.1, and two joining regions, J1.1/4 and J4/2. Nucleotides are numbered from the site of cleavage (black arrow). Dotted lines indicate hydrogen bonding interactions inferred from crystal structures.2,6–8 In the crystallography construct, the binding site for the U1A protein (gray nucleotides) was inserted into the P4 pairing. Black dashed box represents the part of the P4 stem truncated in MD simulations. The site of the C44-G73 to U44-A73 double mutant (DM) is indicated. (b) Tertiary structure of the self-cleaved form of the ribozyme from PDB ID 1CX0,2 generated using VMD.43 Key residues are colored as in panel a.