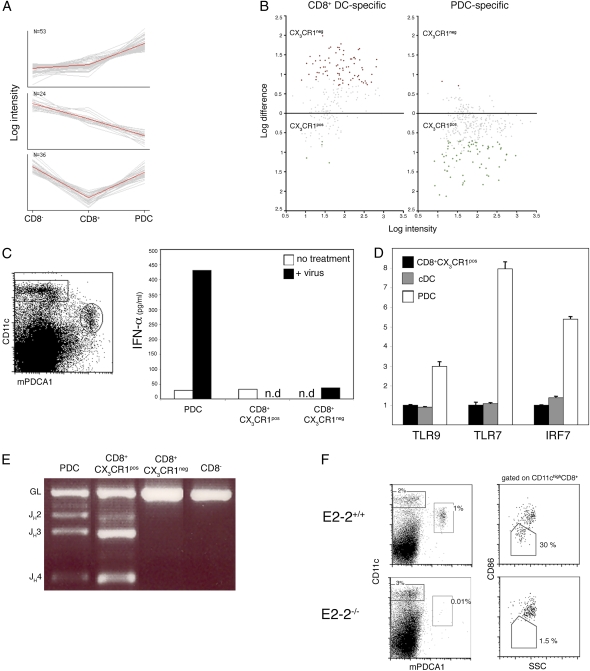
Fig. 4.
CX3CR1+ CD8α+ DC share expression profile, somatic Ig gene rearrangements, and E2-2 dependence with PDC. (A) PCA of the genes overexpressed in CX3CR1+ CD8α+ cells compared with CD8α+ cDC. Shown are the three principal component sets of genes analyzed against the expression data of Robbins et al. (35). Note the preferential expression in PDC (Top), in CD8α− DC (Middle), and in both PDC and CD8α− DC (Bottom). (B) Distribution of genes specific for CD8α+ DC or PDC in the two CD8α+ DC subsets. Shown is pairwise comparison of average probe intensities in CX3CR1+ and CX3CR1− CD8α+ DC; genes specific for CD8α+ DC or PDC were selected based on reference 35. (C) IFN-α production by sorted splenic DC subsets after incubation for 20 h in the presence or absence of 400 HAU/mL influenza virus. (D) Expression of the TLR/IFN signaling genes in sorted CD8α− cDC, PDC, and CX3CR1+ CD8α+ DC as determined by quantitative RT-PCR (mean ± SD of triplicate reactions). Expression levels are normalized relative to the CX3CR1+CD8α+ subset. (E) D–J rearrangement of IgH gene in sorted DC subsets. IgH D–J rearrangements were detected by genomic PCR using primer sets for the DHQ52 element. Data are representative of three experiments. (F) Absence of CX3CR1+ CD8α+ DC and PDC in absence of the transcription factor E2-2. (Left) Gated donor-derived (CD45.2+) splenocytes from E2-2−/− chimeras or control E2-2+/+ chimeras mice were analyzed for the presence of CD11cint Bst2+ PDC. (Right) Gated CD11chi CD8α+ DC comprise the CD86lo SSClo and the CD86hi SSChi subsets (corresponding to CX3CR1+ and CX3CR1− populations, respectively). Data are representative of three independent experiments.