Abstract
The integral peroxisomal membrane proteins PEX10, PEX2, and PEX12 contain a zinc RING finger close to the C terminus. Loss of function of these peroxins causes embryo lethality at the heart stage in Arabidopsis. Preventing the coordination of Zn2+ ions by amino acid substitutions in PEX10, PEX2, and PEX12 and overexpressing the resulting conditional sublethal mutations in WT uncovered additional functions of PEX10. Plants overexpressing ΔZn-mutant PEX10 display deformed peroxisomal shapes causing diminished contact with chloroplasts and possibly with mitochondria. These changes correlated with impaired metabolite transfer and, at high CO2, recoverable defective photorespiration plus dwarfish phenotype. The N-terminal PEX10 domain is critical for peroxisome biogenesis and plant development. A point mutation in the highly conserved TLGEEY motif results in vermiform peroxisome shape without impairing organelle contact. Addition of an N-terminal T7 tag to WT PEX0 resulted in partially recoverable reduced growth and defective inflorescences persisting under high CO2. In contrast, plants overexpressing PEX2-ΔZn-T7 grow like WT in normal atmosphere, contain normal-shaped peroxisomes, but display impaired peroxisomal matrix protein import. PEX12-ΔZn-T7 mutants exhibit unimpaired import of matrix protein and normal-shaped peroxisomes when grown in normal atmosphere. During seed germination, glyoxysomes form a reticulum around the lipid bodies for mobilization of storage oil. The formation of this glyoxysomal reticulum seemed to be impaired in PEX10-ΔZn but not in PEX2-ΔZn-T7 or PEX12-ΔZn-T7 plants. Both cytosolic PEX10 domains seem essential for peroxisome structure but differ in metabolic function, suggesting a role for this plant peroxin in addition to the import of matrix protein via ubiquitination of PEX5.
Keywords: Thr Leu Gly Glu Glu Tyr motif in peroxin PEX10, glyoxysome, metabolomics
Peroxisomes perform multiple metabolic processes, including β-oxidation and H2O2 inactivation by catalase. In plants, specialized peroxisomes, the glyoxysomes, contain glyoxylate cycle enzymes for lipid mobilization. Leaf peroxisomes interact with chloroplasts and mitochondria in photorespiration, a metabolic pathway turning two molecules of glycolate in a series of reactions through glyoxylate, glycine (Gly), serine (Ser), and hydroxypyruvate into CO2 and phosphoglycerate (1 –3). When CO2 is limiting, ribulose-bisphosphate carboxylase/oxygenase functions as an oxygenase and protects the photosynthetic machinery from photodamage by using energy for photorespiration. Mutants lacking enzymes of the photorespiratory cycle are incapable of surviving in ambient air but are able to grow normally in a CO2-enriched atmosphere (2). When genes responsible for plant peroxisome biogenesis are impaired, the phenotypes are severe, ranging from sucrose dependency during early seedling development to embryo lethality (4). The biogenesis of peroxisomes relies on a common class of evolutionary conserved genes referred to as “PEX genes” (5, 6).
The import of peroxisomal matrix protein can be divided into four steps: receptor–cargo interaction, docking at the peroxisome membrane, translocation and cargo release, and receptor recycling. The peroxisomal targeting signals PTS1 and PTS2 are bound by the cytosolic receptors PEX5 and PEX7, respectively, which target the protein to the docking complex (PEX13 and PEX14). The receptors translocate with their cargo into the peroxisome lumen, release the cargo, and are recycled back into the cytosol upon ubiquitination for further rounds of protein import. Ubiquitination requires ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (UBC or E2), and ubiquitin ligase (E3). The RING-finger complex (PEX2, PEX10, PEX12) is required for receptor recycling. PEX10 is linked to the E2 ubiquitin-conjugating enzyme PEX4 (7). The RING-finger peroxins are putative E3 ubiquitin ligases.
PEX10 (At2g26350), PEX2 (At1g79810), and PEX12 (At3g04460) encode integral membrane proteins that share a C3HC4 (HCa)-type zinc-binding RING-finger motif in their C-terminal domain. The PEX10 and PEX2 C3HC4 RING domains bind two Zn2+ ions. PEX12 binds only one Zn2+ ion through its incomplete C5 RING that lacks three of the eight metal ligands (8). The TLGEEY motif (amino acids 91–96) and a highly conserved P motif (amino acid 126) in the N-terminal part of PEX10 are typical of the PEX10 subfamily of zinc-binding proteins (9, 10).
PEX5 in Saccharomyces cerevisiae is modified by polyubiquitination, being dependent on the E2 enzymes Ubc4 and Ubc5. This modification of PEX5 leads to degradation in the 26S proteasome. Monoubiquitination mediated by the E2 enzyme PEX4 results in the recycling of PEX5 for another round of import (11 –13). PEX10 and PEX12 can interact with PEX5 in mammals (14, 15). Impairment of PEX5 ubiquitination and thus of receptor recycling should lead to impaired peroxisomal matrix protein import.
In Arabidopsis, the lack of any of the RING finger peroxins results in embryo lethality (10, 16 –18). We started to elucidate the function of these peroxins using conditional sublethal mutants of PEX10. Peroxisomes can be visualized in transgenic Arabidopsis by the addition of the peroxisomal targeting signal 1 (PTS1) to GFP. A general deficiency in protein import in the peroxisome matrix would lead to mislocalization of GFP-PTS1. We used this technique to analyze the function of the zinc RING finger in PEX10, PEX2, and PEX12.
Results
Characterization of the Mutants Studied.
The conditional sublethal mutant expressing pex10 with a dysfunctional zinc finger in the presence of the WT allele (PEX10-ΔZn1) (19) was crossed with WT plants expressing the peroxisome-localized GFP (GFP-PTS1) (20) to analyze photosynthetic yield and photorespiration in the hybrid plants. Pex10, pex2, and pex12 with a dysfunctional zinc finger (Fig. S1) and an N-terminal T7 tag were transformed into WT GFP-PTS1 plants to determine the photosynthetic and photorespiratory characteristics of PEX10-ΔZn-T7-GFP-PTS1, PEX2-ΔZn-T7-GFP-PTS1, and PEX12-ΔZn-T7-GFP-PTS1 transgenic lines (Fig. 1). The targeting-induced local lesions in genomes (TILLING) line PEX10-W313* with a stop codon 14 amino acids upstream of the zinc finger (Fig. S1) was used as control. We rescued the pex10-KO mutant with the PEX10-WT-T7 cDNA and overexpressed the PEX2-WT-T7 and PEX12-WT-T7 cDNA in WT GFP-PTS1 plants as a control.
Fig. 1.
Phenotype of PEX10, PEX2, and PEX12 mutants grown under 360 or 1,800 ppm CO2. Plants are shown at an age of 28 d (A, D, G, M, and P) and at 46 d (B, C, E, F, H–L, N, O, and Q). (A, D, G, M) (Upper) 360 ppm CO2. (Lower) 1,800 ppm CO2. (B, E, H, I, N, P, Q) 360 ppm CO2. (C, F, K, L, O) 1,800 ppm CO2. (A–F and M-–Q) Control plants GFP-PTS1 WT (Left) and PEX10-ΔZn1 (Right). (A–C) Two PEX10-ΔZn1×GFP-PTS1 hybrids : normal growth only under high CO2, defect in photorespiration. (D–F) Three PEX10-ΔZn-T7-GFP-PTS1 plants: dwarf phenotype under low and high CO2. (G–L) PEX10-WT-T7-pex10-KO plants show dwarf phenotype under low and high CO2. (G Upper and H) Two PEX10-WT-T7-pex10-KO plants (Left) and one WT plant (Right). (G Lower and K) PEX10-WT-T7-pex10-KO (Left), PEX10-ΔZn1 (Center), and WT (Right). (I and L) Mutants have smaller siliques and exhibit a nodding inflorescence in comparison to WT showing bigger siliques and an upright inflorescence. (M–O) One PEX2-ΔZn-T7-GFP-PTS1 and one PEX2-WT-T7-GFP-PTS1 plant grow normally under low and high CO2 with no defect in photosynthetic yield. (P and Q) One WT plant, two PEX12-ΔZn-T7-GFP-PTS1, and one PEX12-WT-T7-GFP-PTS1 transgenic plants grow normally under low and high CO2 with no defect in photosynthetic yield.
The lines were grown in normal air (360 ppm CO2) or under 5-fold elevated CO2 partial pressure for noticeable phenotypes that could be rescued by high CO2. GFP-PTS1 WT plants and the PEX10-ΔZn1 line showing dwarfism in normal air and rescue under high CO2 were used as controls (Fig. 1).
The PEX10-ΔZn1×GFP-PTS1 hybrid had a chlorotic, photorespiratory phenotype (Fig. 1 A–C) and reduced photosynthetic yield (Fig. S2A) in normal air that was rescued by elevated CO2. Three PEX10-ΔZn-T7-GFP-PTS1 transgenic lines displayed dwarfism in normal air, especially while flowering. Only plant height was reduced in these lines, and growth under high CO2 rescued lines 2 and 3 to some extent, but not line 1 (Fig. 1 D–F); their photosynthetic yield reached nearly WT level in normal air and high CO2 (Fig. S2A). Overexpression of the PEX10-WT-T7 cDNA in the pex10-KO mutant rescued the embryo-lethal phenotype, as did PEX10-WT cDNA (10). Rescued plants at flowering stage had a dwarfish phenotype in normal air that was rescued only slightly at high CO2; they had shorter internodes, a nodding inflorescence, and smaller siliques than WT (Fig. 1 G–L). The dwarfish phenotype caused by the N-terminal T7 tag seems epistatic to growth retardation in PEX10-ΔZn plants that can be rescued by high CO2.
Lines homozygous for pex2-ΔZn-T7 cDNA or for PEX2-WT-T7 cDNA in the WT GFP-PTS1 background had equal growth in normal air or under high CO2; overexpressing the mutated pex2-ΔZn-T7 or the PEX2-WT-T7 did not elicit growth retardation, a photorespiratory phenotype, or chlorosis (Fig. 1 M–O). They had WT photosynthetic yield in normal air and under high CO2 (three independent pex2-ΔZn-T7 lines are shown in Fig. S2C). Growth of the PEX12-ΔZn-T7-GFP-PTS1 lines and the PEX12-WT-T7-GFP-PTS1 lines in normal air did not differ from the growth of WT (Fig. 1 P–Q). Their photosynthetic yield was similar to that of WT (Fig. S2D).
PEX10 exhibits two highly conserved motifs: TLGEEY (amino acids 91–96) and 126P (Fig. S1). We crossed the PEX10-TILLING lines to WT GFP-PTS1 plants. The changes in amino acids cause various degrees of stunting (Fig. S3A), but the photorespiratory reaction was retained (Fig. S2B).
Thus three groups of mutants could be distinguished: (i) mutants (GFP-PTS1, PEX2-ΔZn-T7-GFP-PTS1, PEX2-WT-T7-GFP-PTS1, PEX12-ΔZn-T7-GFP-PTS1, and PEX12-WT-T7-GFP-PTS1) that, like WT, grow normally under ± CO2, with no defect in photosynthetic yield; (ii) a mutant (PEX10-ΔZn1×GFP-PTS1) that grows normally only under +CO2 and has a defect in photosynthetic yield only under −CO2 (the plant has a defect in photorespiration); (iii) mutants (PEX10-ΔZn-T7-GFP-PTS1, PEX10-W313*, PEX10-G93E, PEX10-P126S, and PEX10-WT-T7-pex10-KO) that show dwarf phenotype under ±CO2 but no defect in photosynthetic yield (these plants have defects not related to photorespiration).
Transcript levels of the endogenous WT genes and the mutated pex10, pex2, and pex12 transgenes were determined by quantitative RT-PCR (qRT-PCR) (Fig. S4 A–C). The three PEX10-ΔZn-T7-GFP-PTS1 transgenic lines had 110-fold (line I), 10-fold (line II), or 65-fold (line III) transcription levels relative to WT (Fig. S4A). In the PEX10-ΔZn1×GFP-PTS1 hybrids, transcription of both the WT and pex10-ΔZn gene together was much lower (Fig. S4A) with approximately equal amounts of WT and mutated transcript (19). The PEX10-WT-T7 cDNA in the pex10-KO mutant, which lacks the endogenous WT gene, exhibited a 60-fold higher transcription rate relative to WT (PEX10-WT-T7-pex10-KO; Fig. S4 A and E). The three PEX2-ΔZn-T7-GFP-PTS1 transgenic lines exhibited a 50-fold (line I), 150-fold (line II), or 40-fold (line III) transcription level relative to WT (Fig. S4B). The two PEX12-ΔZn-T7-GFP-PTS1 transgenic lines exhibited a 5-fold (line II) or 8-fold (line IV) transcription level relative to WT (Fig. S4C). These results were confirmed by determining transcript levels by qRT-PCR (Fig. S4 D, F, and G).
Overexpression of PEX10 with a Dysfunctional Zinc RING Finger in the WT Background Correlates with Abnormal Peroxisome Morphology and Loss of Contact Between Chloroplasts and Leaf Peroxisomes Without Impairment of Peroxisomal Matrix Protein Import.
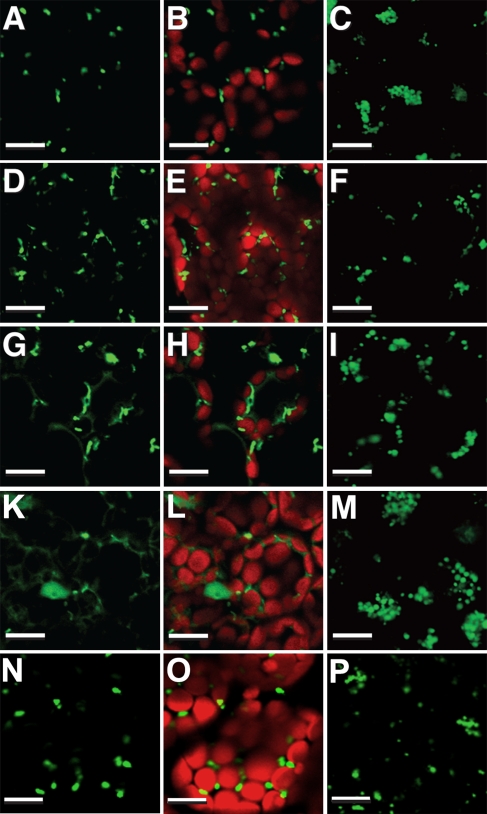
Leaf-type peroxisomes in 14-d-old secondary leaves, as well as glyoxysomes in 5-d-old dark-grown cotyledons in PEX10-ΔZn1×GFP-PTS1 hybrids and in PEX10-ΔZn-T7-GFP-PTS1 transgenics, were examined by confocal laser scanning microscopy (CLSM) (Fig. 2). GFP-PTS1 WT plants showed punctuate leaf peroxisomes mainly in close contact with chloroplasts (Fig. 2 A and B). The leaf peroxisomes in PEX10-ΔZn hybrids (Fig. 2 D and E) and in PEX10-ΔZn-T7 transgenics (Fig. 2 G and H) were pleiomorphic and rarely associated with chloroplasts. The formation of a glyoxysomal reticulum was well elaborated, if highly variable, in WT plants (Fig. 2C) but not in PEX10-ΔZn hybrids or PEX10-ΔZn-T7 transgenics (Fig. 2 F and I). The PEX10-ΔZn1×GFP-PTS1 hybrid did not show mislocalization of GFP-PTS1 (Fig. 2 D–F). In the PEX10-ΔZn-T7-GFP-PTS1 transgenic (Fig.2 G–I), a barely visible impairment of matrix protein importation was recognizable around leaf peroxisomes.
Fig. 2.
Peroxisome morphology and importation of matrix protein in PEX10-ΔZnxGFP-PTS1 hybrids and in PEX10-ΔZn-T7-GFP-PTS1, PEX2-ΔZn-T7-GFP-PTS1, and PEX12-ΔZn-T7-GFP-PTS1 transgenics visualized by GFP-PTS1. (Left) GFP signal from leaf peroxisomes in 14-d-old secondary leaves. (Center) GFP signal merged with chloroplast autofluorescence. (Right) GFP signal from glyoxysomal reticulum of 5-d-old dark-grown cotyledons. (A–C) GFP-PTS1 WT. (D–E) PEX10-ΔZn1xGFP-PTS1 hybrid. (G–I) PEX10-ΔZn-T7-GFP-PTS1 transgenic. (K–M) PEX2-ΔZn-T7-GFP-PTS1 transgenic. (N–P) PEX12-ΔZn-T7-GFP-PTS1 transgenic. (Scale bar, 10 μm.)
PEX10-W313* TILLING lines with a stop codon in front of the C3HC4-zinc RING finger were analyzed. The point mutation was verified by PCR. Heterozygous plants have a 1:1 ratio of intact and mutated PEX10. Loss of the zinc finger leads, as expected, to embryo lethality in homozygous plants. Heterozygous PEX10-W313* plants were not chlorotic but had a dwarfish phenotype in normal air that could be rescued to some extent under high CO2 (Fig. S3A); the photosynthetic yield was similar to that of WT in normal air and high CO2 (Fig. S2B). Peroxisomes in Arabidopsis WT mesophyll cells were spherical or ovoid, appressed to the envelope of chloroplasts, and frequently in close physical contact with mitochondria (Fig. S5 A and B), whereas PEX10-W313* cells showed misshapen peroxisomes without contact with other organelles. In WT cells, 88 ± 16% of peroxisomes were appressed to a chloroplast, whereas in PEX10-W313* cells 24 ± 34% were appressed to chloroplasts (P < 0.01, n = 10) (Fig. S5 G and H). PEX10-W313* plants thus were similar to PEX10-ΔZn plants.
Morphological analysis disclosed two peroxisomal populations in PEX10-ΔZn plants: WT (mostly spherical) and elongated, pleiomorphic organelles with significantly reduced contact with chloroplasts (Table 1). Examination of living cells with fluorescently labeled peroxisomes revealed that those peroxisomes move independently of chloroplasts (Movie S1 shows WT plants; Movie S2 shows PEX10-ΔZn plants).
Table 1.
Peroxisome abundance and form in WT and PEX10 mutants
| Peroxisome abundance and form | WT | PEX10-G93E | PEX10-ΔZn1 |
| No. peroxisomes/cell section (n = 25 cells) | 3.3 ± 1.5 | 3.0 ±1.6 | 9.0 ± 4.2 |
| Percent peroxisomes attached to chloroplasts (n = 25 cells) | 88 | 91 | 34 |
| Percent peroxisomes with normal, round shape (formfactor >0.732) (n = 105) | 89 | 62 | 77 |
| Percent peroxisomes with deformed, worm-like shape(formfactor <0.732) (n = 105) | 11 | 38 | 23 |
Overexpression of PEX2 with a Dysfunctional Zinc RING Finger in the WT Background Results in Impaired Peroxisomal Matrix Protein Import with Normal Peroxisome Morphology.
CLSM of PEX2-ΔZn-T7-GFP-PTS1 transgenics revealed significantly impaired import of matrix protein in leaf peroxisomes, as shown by mislocalization of GFP-PTS1, although leaf peroxisomes were normal (Fig. 2 K and L). Mislocalization of GFP-PTS1 also was seen in the glyoxysomes of dark-grown cotyledons, although the glyoxysomal reticulum was normal (Fig. 2M). Plants overexpressing the PEX2-WT-T7 cDNA had glyoxysomes with normal morphology and leaf peroxisomes without mislocalization of GFP-PTS1. The relative activity of serine glyoxylate aminotransferase in peroxisomes of the PEX2-ΔZn-T7-GFP-PTS1 transgenic lines was 40% and the relative activity of hydroxypyruvate reductase was 80% of WT and PEX2-WT-T7-GFP-PTS1 lines. Peroxisome formation and contact with chloroplasts was similar in WT (Fig. S5 A and B) and PEX2-ΔZn-T7 transgenics (Fig. S5 I and K). Thus, PEX2 is involved in matrix protein import, but a certain decrease in the level of matrix enzymes causes no defect in photorespiration.
Overexpression of PEX12 with a Dysfunctional Zinc RING Finger in the WT Background Has No Effect on Peroxisome Formation or Matrix Protein Import.
No differences between mutated and WT plants were seen in PEX12-ΔZn-T7-GFP-PTS1 transgenics homozygous for the pex12-ΔZn-T7 construct. Growth and morphology of leaf peroxisomes and glyoxysomes were similar to WT, and there was no mislocalization of GFP-PTS1 (Fig. 2 N–P). Dilution of the endogenous PEX12 by the PEX12-ΔZn-T7 had no effect on matrix protein import or peroxisome formation. PEX12-WT-T7-GFP-PTS1 transgenics also exhibit normal shaped peroxisomes and no import deficiency.
PEX10 Contains Two Essential Motifs for Peroxisome Biogenesis in the N-Terminal Region.
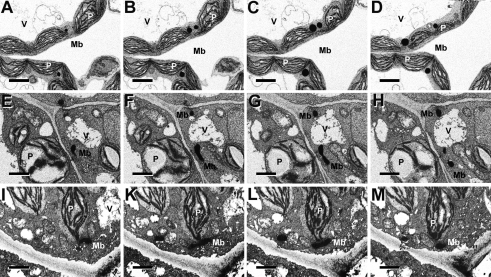
The PEX10-G93E and PEX10-P126S TILLING lines were crossed twice to WT GFP-PTS1. PEX10-G93E×GFP-PTS1 hybrids produced homozygous seeds that gave rise to seedlings that could not survive in the light, but there was no difference from WT when grown for 14 d in the dark (Fig. S3 B and D). Heterozygous PEX10-G93E lines had a dwarfish phenotype in normal air that could be rescued to some extent in high CO2 (Fig. S3A). Homozygous PEX10-P126S exhibited growth retardation in normal air and high CO2 in the light (Fig. S3A) but not in the dark (Fig. S3 C and E). Neither line was chlorotic. Their photosynthetic yield was similar in normal air and high CO2 (Fig. S2B). When analyzed by CLSM, PEX10-G93E×GFP-PTS1 hybrids revealed deformed worm-like clustered peroxisomes; PEX10-P126S×GFP-PTS1 hybrids had normal peroxisomes; in both cases, no GFP signal could be detected in the cytosol, indicating unimpaired matrix protein import (Fig. S3). The morphology and association of leaf peroxisomes with other organelles was examined by light microscopy, transmission electron microscopy (Fig. S5), and focused ion beam (FIB) electron microscopy (Fig. 3). PEX10-G93E plants had elongated vermiform peroxisomes, but they were appressed to chloroplasts (91%, n = 25 cells). The mutation of the TLGEEY motif in the N terminus had an effect on peroxisome formation similar to that of overexpression of the PEX10-ΔZn without affecting organelle contact (Fig. S5 C and D). In homozygous PEX10-P126S plants, peroxisomes were slightly smaller and often grouped compared with WT, but contact between peroxisomes and chloroplasts was not impaired (Fig. S5 E and F). Serial FIB electron micrographs verified the different morphologies and chloroplast-association patterns in WT, PEX10-ΔZn, and PEX10-G93E plants (Fig. 3).
Fig. 3.
Focused ion beam (FIB) electron micrographs of 13-d-old leaf tissue of WT (Top row), PEX10-ΔZn1 (Middle row) and PEX10-G93E (Bottom row). The tissue was stained for catalase activity with diaminobenzidine. Images at successive levels of sections exposed by the ion beam are shown. (A–D) Leaf peroxisomes of WT plants are ovoid and in physical contact with chloroplasts. (E–H) Leaf peroxisomes of PEX10-ΔZn1 are worm-like and accumulate at places distant from chloroplasts. (I–M) Leaf peroxisomes of PEX10-G93E plants are elongated but are attached to chloroplasts. Mb, microbody (peroxisome); P, chloroplasts; V, vacuole. (Scale bar, 3 μm.)
Morphological analysis revealed two peroxisomal populations in PEX10-G93E plants (n = 105 peroxisomes): WT (spherical peroxisomes) and deformed worm-like peroxisomes that had contact with chloroplasts as in WT (Table 1).
Metabolome Analysis in PEX10-ΔZn Plants Confirms Impaired Organelle Contact or Impairment of the Glycolate Cycle.
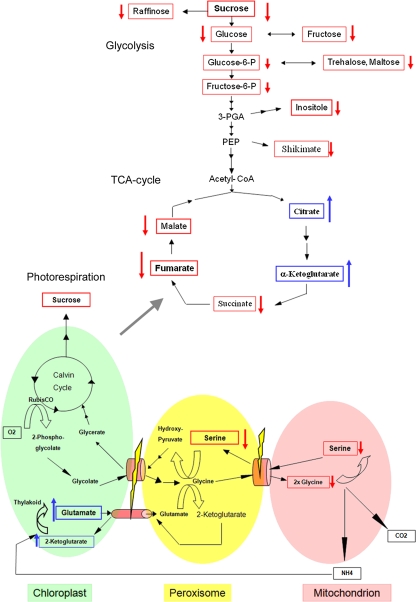
WT and PEX10-ΔZn1 plants were grown for 14 d in normal air either in the greenhouse (Fig. S6A Upper) or in phytochambers (Fig. S6A Lower). Metabolome analysis was carried out on 10- to 15-mg leaf samples extracted and supplied with the necessary reagents (SI Text). WT and PEX10-ΔZn1 plants showed different metabolite abundances by principal component analysis (PCA) (Fig. S6A), and 22 metabolites (including the photorespiratory metabolites Gly, Ser, and glycerate) had significantly different concentrations (Fig. S6B). Gly, Ser, and glycerate were decreased, and Glu was increased in PEX10-ΔZn1 plants compared with WT (Fig. 4, Fig. S6C, and Fig. S7A).
Fig. 4.
Model of the photorespiration pathway with link to TCA cycle and glycolysis for the dysfunctional metabolic flux for PEX10-ΔZn1 mutants compared with WT plants. Identified significant reductions or accumulations are marked with red (reduced) or blue (accumulated) arrows. The localization of the measured metabolites is postulated because of the metabolic profiling performed without regard to compartmentalization of the cell.
Loss of Contact Between Peroxisomes and Chloroplasts in PEX10-ΔZn Plants Is Not Caused by Oxidative Stress.
WT and PEX10-ΔZn1 plants were grown for 14 d in phytochambers under elevated CO2 where growth retardation and chlorosis stress symptoms were absent. PCA showed a clear separation of WT and PEX10-ΔZn1 samples (Fig. S8A). The glycolate cycle intermediates Gly, Ser, and glycerate had generally lower concentrations, probably because of the reduction of oxygenase activity (Fig. S8 B and C), but still were significantly reduced in PEX10-ΔZn1 compared with WT, suggesting that a disturbed metabolite flow is caused by impaired organelle contact (Fig. S7B) rather than by oxidative stress resulting from impaired photorespiration. Contact between peroxisomes and chloroplasts seemed to be impaired even without oxidative stress, suggesting that the dysfunctional zinc RING finger in PEX10 leads to loss of organelle contact.
Metabolome Analysis of PEX10-G93E and PEX10-P126S Mutant Plants Confirms an Unimpaired Functional Contact Between Peroxisomes and Chloroplasts Despite Altered Peroxisome Morphology.
The PEX10-TILLING lines had metabolite profiles different from both WT and PEX2-ΔZn-T7 (Fig. S9A). Gly, Ser, and glycerate were significantly elevated in PEX10-G93E plants as compared with WT, whereas Ser, Glu, and alanine (Ala) were significantly elevated in PEX10-P126S plants (Fig. S9 B and C). The PEX10-TILLING lines accumulate Ser, but behave differently otherwise. As a consequence, the Ser/glycerate ratio is elevated for both PEX10-TILLING lines (Fig. S7C). The flow of metabolites within the glycolate cycle in PEX10-TILLING lines is not impaired, suggesting that neither the aberrant peroxisome form of PEX10-G93E plants nor the small and assembled peroxisomes in PEX10-P126S plants led to a reduction of photorespiratory metabolites.
Metabolome Analysis Confirms That the Glycolate Cycle in PEX2-ΔZn-T7 Plants Is Not Impaired Despite Reduced Import of Matrix Protein.
PEX2-ΔZn-T7 and WT are not distinguished by metabolite variances, indicating a minor effect on metabolite profiles (Fig. S9D). The t-test clustering shows metabolites differing significantly in PEX2-ΔZn-T7 and WT (Fig. S9E). Mutant plants exhibited elevated values for the glycolate cycle intermediates Ser, glycerate, and Ala (Fig. S9F), but with low statistical significance, and the Ser/glycerate ratio is similar to that in WT (Fig. S7D). The impaired peroxisomal matrix protein import in PEX2-ΔZn-T7 plants did not influence the flow of photorespiratory metabolites between peroxisomes and chloroplasts.
Discussion
The partial loss-of-function PEX10-ΔZn mutants exhibited normal peroxisomal matrix protein import but formed multilobed peroxisomes that are not appressed to the chloroplast. The levels of markers for the C2 cycle of the photosynthetic carbon metabolism (Gly, Ser, and glycerate) (3, 21) were significantly reduced, and Glu accumulated, probably in chloroplasts. The 30% reduction of the Ser/glycerate ratio probably was caused by a block in glycolate flowing from chloroplasts to peroxisomes in the mutants, resulting in a reduced supply of Ser via the C2 cycle (Fig. 4). Unlike similar morphological changes resulting from exposure to hydrogen peroxide or hydroxyl radicals (22), oxidative stress could be ruled out as the reason for the loss of organelle contact in PEX10-ΔZn mutants, because plants grown under elevated CO2 exhibited WT levels of growth and no indication for oxidative stress. Accumulation of Glu and the reduced Ser/glycerate ratio indicate a disturbed metabolite flux caused by reduced organelle contact, which is expressed as metabolic phenotype under normal and high CO2 conditions. Tricarboxylic acid (TCA) cycle and glycolysis intermediates also are changed significantly and could indicate a modified energy state or a global effect on primary metabolism. Lower photorespiratory flux rates require less NADH for hydroxypyruvate reduction; NADH is provided to the peroxisome by the oxidation of malate from chloroplasts and mitochondria, and intraperoxisomal NADH generation is seen as the rate-limiting step in the conversion of Ser to glycerate (3).
Mutating the N-terminal TLGEEY motif causes pleiomorphic peroxisomes that retain contact between peroxisomes and chloroplasts; the matrix protein import is undisturbed, but photorespiratory marker levels Gly, Ser, and glycerate are increased, as is the Ser/glycerate ratio. Accumulation of Gly and Ser occurs in the mutants with reduced hydroxypyruvate reductase activity, whereas increased glycerate indicates reduced phosphoglycerate kinase (21). Mutant PEX10-P126S has normal peroxisomal protein import and organelle contact but accumulates Ser, whereas Gly and glycerate levels are unaffected. These different specific effects, as well as the disturbances of adding a T7 tag to PEX10-WT and to PEX10-ΔZn, may indicate that the organelle contact is impaired for specific metabolite transfer.
In PEX2-ΔZn-T7 plants the import of peroxisome proteins was reduced to 40% of WT, while peroxisome morphology and contact to chloroplasts were normal, but Ser and glycerate levels were increased. PEX2-WT-T7, PEX12-WT-T7, and PEX12-ΔZn-T7 plants exhibit no impairment of matrix protein import, peroxisomal morphology, or metabolic flow.
The RING peroxins PEX10, PEX2, and PEX12 are thought to function in a complex in matrix protein import by ubiquitinating the receptor (1). ScPEX10 acts as the E3 ligase for Ubc4-dependent ubiquitination of PEX5 but not for PEX4-dependent ubiquitination (23). In Arabidopsis, pex12 mutants are defective in protein transport to peroxisomes, as shown by mislocalization of GFP-PTS1 (24). These data indicate ubiquitination requires not only separate E2 enzymes but also separate E3 ligases.
We found aberrant peroxisome morphology in PEX10-ΔZn, PEX10-ΔZn-T7, and PEX10-G93E lines but no import deficiency, whereas PEX2-ΔZn-T7 lines exhibit only import deficiency. The import deficiency in some PEX10-ΔZn-T7 lines might be caused by instability of the RING complex resulting from the altered N terminus with the T7 tag; this import deficiency, however, was much less pronounced than in the PEX2-ΔZn-T7 lines. PEX12-ΔZn-T7 lines showed no phenotype.
Given the embryo lethality of the peroxin KO mutants, down-regulation of PEX gene expression using RNA interference (knockdown mutants) is another way to determine PEX10, PEX2, and PEX12 function (25). The pex10i mutant showed pleiotropic phenotypes such as reduced cell size and variegated leaves, which were not observed in other pex mutants. Reduced expression of 38% (pex10i), 31% (pex2i), and 25% (pex12i) could be achieved. Misdistribution of peroxisomal matrix proteins but no aberrant peroxisomal morphology was observed in all cases (20). These results differ from those achieved by our approach, in which a 1:1 dilution of the intact endogenous PEX10 with the dysfunctional PEX10-ΔZn resulted in 50% activity. Because peroxisomes of the pex10i line show normal peroxisomal morphology, whereas PEX10-ΔZn mutants show aberrant peroxisomal morphology but no import deficiency, we conclude that the deficiency in peroxisomal protein import in RNAi lines is caused by reduced complex formation between PEX10, PEX2, and PEX12. PEX10 cannot be involved directly in ubiquitination of PEX5 for recycling to the cytosol, because PEX10-ΔZn plants do not exhibit impaired matrix protein import. We hypothesize that PEX10 is necessary for the stability of the RING complex, whereas PEX2 is responsible for the monoubiquitination of PEX5.
PEX10 is necessary for normal peroxisome morphology. Mutations in the TLGEEY motif of the N terminus also impair peroxisome morphology but do not significantly reduce contact with the chloroplast envelope. The peroxisomes seen in the mutant plant cells may be the result of an increase in membrane surface area, a decrease in volume, or a combination of both. These observations emphasize the general importance of PEX10 in peroxisome morphology and of the PEX10 zinc RING finger for organelle contact. In contrast, PEX2 is important for the matrix protein import and does not seem to be involved in determining peroxisome morphology or organelle contact. During germination, glyoxysomes build a reticulum around lipid bodies for storage oil mobilization. The formation of this glyoxysomal reticulum, which is impaired in PEX10-ΔZn mutants but not in PEX2-ΔZn and PEX12-ΔZn plants, supports the role of PEX10 as an early peroxin (10).
The involvement of PEX10 in organelle contact and its possible connection with metabolite transfer, especially in photorespiration, is unique among the RING peroxins and might be specific for plants.
Methods
Generation of PEX10, PEX2, and PEX12 Plants with Dysfunctional Zinc RING Finger (PEX10-ΔZn, PEX2-ΔZn, PEX12-ΔZn) in the GFP-PTS1 WT Background.
The PEX10-ΔZn1 line of our former analysis (19) was crossed with Arabidopsis Columbia WT plants expressing GFP-PTS1 (20), resulting in PEX10-ΔZn1×GFP-PTS1 plants; pex10-ΔZn-T7, pex2-ΔZn-T7, and pex12-ΔZn-T7 cDNAs were constructed with an N-terminal T7 tag and a triple Gly-Ser linker between the T7 tag and the start codon and were transformed into Arabidopsis Columbia GFP-PTS1 WT plants (20). The relative expression level of endogenous and transgenic PEX genes was determined by qRT-PCR (SI Text).
Genomic Verification of PEX10-G93E, PEX10-P126S, and PEX10-W313* TILLING Lines and Crossing into the GFP-PTS1 WT Background.
For mutant background cleaning and to introduce GFP staining of peroxisomes, we crossed the PEX10-G93E and PEX10-P126S TILLING lines twice to WT GFP-PTS1 (20). Point mutations were verified by PCR. Details are given in SI Text.
Plant Growth Conditions.
Plants were grown under long-day conditions in normal atmosphere (360 ppm CO2) or high CO2 (1,800 ppm CO2) adjusted in an open gas exchange system including phytochambers (SI Text).
Enzyme Assays and Photosynthetic Parameters.
Enzyme assays and photosynthetic parameters were estimated as described (19) (SI Text).
Confocal Microscopy.
Primary leaves of 3-wk-old plants were analyzed on Olympus FluoView FV1000 (Olympus) (SI Text).
Electron and Light Microscopy and Cytochemical Staining.
Electron and light microscopy and cytochemical staining for catalase were performed as described (19) (SI Text).
Metabolome Analysis.
Homogenized plant samples were analyzed by gas chromatography coupled to a LECO Pegasus IV TOF mass analyzer (SI Text).
Statistical Analysis.
Univariate and multivariate data analysis was done with The Institute for Genomic Research Multi Experiment Viewer (TIGR MeV; OpenSource Software, http://www.tm4.org/mev/), version 4.1 (SI Text).
Supplementary Material
Acknowledgments
We thank M. Nishimura and M. Hayashi (National Institute for Basic Biology, Okazaki, Japan) for Arabidopsis seeds expressing GFP-PTS1 and H. Schnyder (Technische Universität München, Germany) for the use of phytochambers with an open gas exchange system. We thank the Arabidopsis Biological Research Center (ABRC), the Nottingham Stock Centre (NASC), the RIKEN Biological Resource Center Experimental Plant Division, and the Institut National de Recherche Agronomique (INRA) for providing material. This work was supported by Deutsche Forschungsgemeinschaft Grant Gi 154/12-2.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009174107/-/DCSupplemental.
References
- 1.Brown LA, Baker A. Shuttles and cycles: Transport of proteins into the peroxisome matrix (review) Mol Membr Biol. 2008;25:363–375. doi: 10.1080/09687680802130583. [DOI] [PubMed] [Google Scholar]
- 2.Somerville CR. An early Arabidopsis demonstration. Resolving a few issues concerning photorespiration. Plant Physiol. 2001;125:20–24. doi: 10.1104/pp.125.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reumann S, Weber AP. Plant peroxisomes respire in the light: Some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim Biophys Acta. 2006;1763:1496–1510. doi: 10.1016/j.bbamcr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Hayashi M, Nishimura M. Arabidopsis thaliana—a model organism to study plant peroxisomes. Biochim Biophys Acta. 2006;1763:1382–1391. doi: 10.1016/j.bbamcr.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Mano S, Nishimura M. Plant peroxisomes. Vitam Horm. 2005;72:111–154. doi: 10.1016/S0083-6729(05)72004-5. [DOI] [PubMed] [Google Scholar]
- 6.Eckert JH, Erdmann R. Peroxisome biogenesis. Rev Physiol Biochem Pharmacol. 2003;147:75–121. doi: 10.1007/s10254-003-0007-z. [DOI] [PubMed] [Google Scholar]
- 7.Eckert JH, Johnsson N. Pex10p links the ubiquitin conjugating enzyme Pex4p to the protein import machinery of the peroxisome. J Cell Sci. 2003;116:3623–3634. doi: 10.1242/jcs.00678. [DOI] [PubMed] [Google Scholar]
- 8.Stone SL, et al. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren DS, Morrell JC, Moser HW, Valle D, Gould SJ. Identification of PEX10, the gene defective in complementation group 7 of the peroxisome-biogenesis disorders. Am J Hum Genet. 1998;63:347–359. doi: 10.1086/301963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C. AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA. 2003;100:9626–9631. doi: 10.1073/pnas.1633697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiel JAK, Emmrich K, Meyer HE, Kunau W-H. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem. 2005;280:1921–1930. doi: 10.1074/jbc.M403632200. [DOI] [PubMed] [Google Scholar]
- 12.Kragt A, Voorn-Brouwer T, van den Berg M, Distel B. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J Biol Chem. 2005;280:7867–7874. doi: 10.1074/jbc.M413553200. [DOI] [PubMed] [Google Scholar]
- 13.Platta HW, et al. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol. 2007;177:197–204. doi: 10.1083/jcb.200611012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang CC, Warren DS, Sacksteder KA, Gould SJ. PEX12 interacts with PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J Cell Biol. 1999;147:761–774. doi: 10.1083/jcb.147.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okumoto K, Abe I, Fujiki Y. Molecular anatomy of the peroxin Pex12p: Ring finger domain is essential for Pex12p function and interacts with the peroxisome-targeting signal type 1-receptor Pex5p and a ring peroxin, Pex10p. J Biol Chem. 2000;275:25700–25710. doi: 10.1074/jbc.M003303200. [DOI] [PubMed] [Google Scholar]
- 16.Hu JP, et al. A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science. 2002;297:405–409. doi: 10.1126/science.1073633. [DOI] [PubMed] [Google Scholar]
- 17.Sparkes IA, et al. An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 2003;133:1809–1819. doi: 10.1104/pp.103.031252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan JL, et al. The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 2005;139:231–239. doi: 10.1104/pp.105.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumann U, et al. Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc Natl Acad Sci USA. 2007;104:1069–1074. doi: 10.1073/pnas.0610402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mano S, et al. Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: Dynamic morphology and actin-dependent movement. Plant Cell Physiol. 2002;43:331–341. doi: 10.1093/pcp/pcf037. [DOI] [PubMed] [Google Scholar]
- 21.Timm S, et al. A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis . Plant Cell. 2008;20:2848–2859. doi: 10.1105/tpc.108.062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinclair AM, Trobacher CP, Mathur N, Greenwood JS, Mathur J. Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J. 2009;59:231–242. doi: 10.1111/j.1365-313X.2009.03863.x. [DOI] [PubMed] [Google Scholar]
- 23.Williams C, van den Berg M, Geers E, Distel B. Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun. 2008;374:620–624. doi: 10.1016/j.bbrc.2008.07.054. [DOI] [PubMed] [Google Scholar]
- 24.Mano S, Nakamori C, Nito K, Kondo M, Nishimura M. The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 2006;47:604–618. doi: 10.1111/j.1365-313X.2006.02809.x. [DOI] [PubMed] [Google Scholar]
- 25.Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M. Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 2007;48:763–774. doi: 10.1093/pcp/pcm053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.