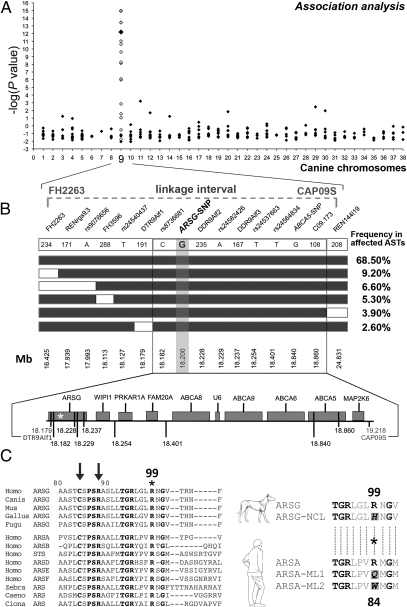
Fig. 2.
Mapping, fine-mapping, and identification of a candidate gene for NCL in ASTs. (A) A single locus with strong genome-wide significance was identified on CFA09 (larger black diamonds) and confirmed (white diamonds); see Table S2. The −log(P values) are reported on the y-axis. The 2.8-Mb candidate region defined by our linkage analysis (18) is shown. (B) Fine-mapping using haplotype analysis for NCL. Haplotypes identified in affected ASTs are shown as boxes (NCL allele, black boxes; alternative allele, white boxes). Genotyped microsatellites and SNPs are indicated in their 5′-3′ position, and their position on CFA09 is shown. The Haplotypes with a frequency <2% are omitted. In the bottom part, candidate genes within the critical region are shown. The relative positions of markers are indicated by vertical black lines, and the nonsynonymous associated ARSG-SNP is represented by an asterisk. (C) (Left) N-terminal sequence of ARSG or predicted ARS from metazoans is aligned with the N-terminal amino acids of human ARS. Conserved amino acids are in bold type. Arrows point to two of the 10 residues involved in the catalytic site of arylsulfatases (22), with positions referring to the canine ARSG. The R99 mutant in affected dogs is highlighted by an asterisk. (Right) The sequence flanking the R99 of the canine WT ARSG protein (ARSG) is aligned with the corresponding sequence of the mutated protein (ARSG-NCL) and best aligned with the R84 of the human WT ARSA (ARSA), replaced by a glutamine (Q) or a tryptophane (W) in some patients affected by metachromatic leukodystrophy [ARSA-ML1; (25) and ARSA-ML2, (24)].