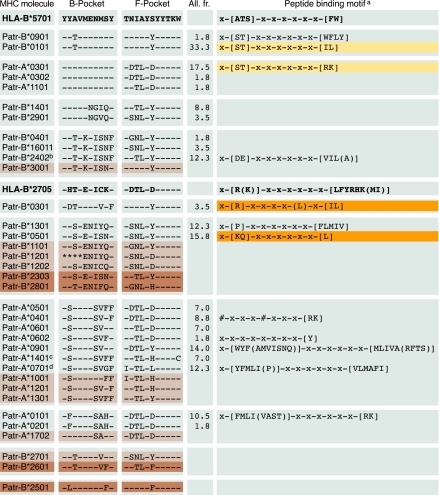
Fig. 1.
Chimpanzee class I molecules clustered based on B- and F-pocket similarities. The B and F pocket of HLA-B*5701 are taken as consensus. A dash indicates identity to the consensus, an amino acid replacement is represented by the conventional one-letter code, and an asterisk indicates the position where the amino acid is unknown. The allele frequency (All. fr.) figures of the wild-caught founder population have been provided. Molecules depicted in light and dark brown are from chimpanzees of the Central and East African subspecies, respectively. Dark orange represents unreported peptide-binding motifs, whereas light orange highlights the motifs that are in agreement with data reported by another research team (23, 24). The binding motifs for HLA-B*2705 and -B*5701 as well as previously identified Patr-binding motifs are provided (23–25, 43). aThe conventional one-letter code identifies the preferred, or in brackets the tolerated, residues at the corresponding positions. bPatr-B*2402 has an identical B and a similar F pocket (–TL-F—–) as Patr-B*2401. Binding motif shown is from Patr-B*2401. cMolecule present in West and East African subspecies. dMolecule present in West, Central, and East African subspecies. The # marks additional primary anchors.