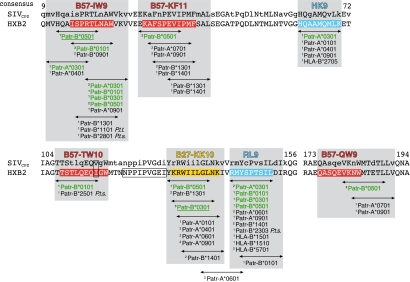
Fig. 2.
Map of potential HIV-1 Gag CTL epitopes recognized by Patr class I molecules. The Gag sequence of HXB2 is taken as consensus, with the consensus of SIVcpz above (http://www.hiv.lanl.gov). A lowercase letter in the SIVcpz consensus indicates a variable position. HLA-B*27 and -B*57 CTL epitopes are marked in yellow and red, respectively. Potential 9-mer epitopes are indicated by an arrow, with the relevant Patr molecule(s) mentioned. Molecules marked in green indicate the predicted MHC/peptide combinations that are tested in peptide-binding studies. Molecules specific for Central (P.t. troglodytes, P.t.t.) or East African chimpanzees (P.t. schweinfurthii, P.t.s.) are indicated. The predicted binding affinities using the NetMHCpan 2.0 algorithm are indicated by 1high, 2intermediate, or 3low. The arrows above the underscored Patr molecules indicate previously reported CTL epitopes (30). The two potential epitopes, HK9 and RL9, identified with the NetMHCpan 2.0 algorithm are marked in blue. The control peptide, NPPIPVGEI, is boxed. The box in the B57-TW10 epitope indicates covered variation (Fig. S2).