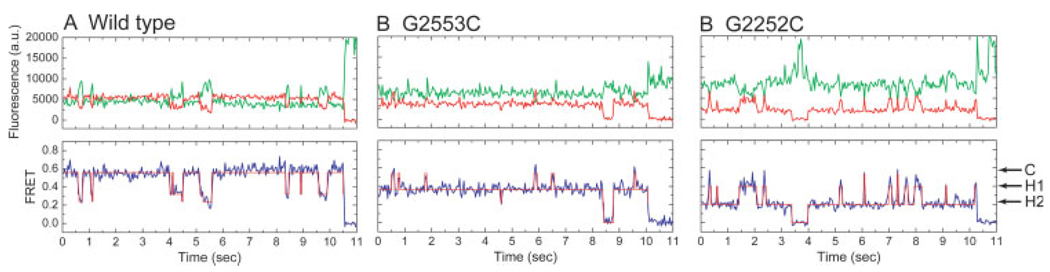
FIGURE 3.
Wide-field, single-molecule FRET measurements provide direct observations of conformational dynamics of tRNAs on individual ribosomes. Anticorrelated changes in donor (green) and acceptor (red) fluorescence of dyes linked to tRNA report on the dynamics of tRNA within the ribosome. The efficiency of FRET (blue) reports on the distance separating the two dyes. Hidden Markov modeling, resulting in the idealization of the FRET trajectories (overlaid in red), is used to identify the FRET value and lifetimes of each FRET state observed as well as the order of transitions in the system. Here, FRET trajectories have been idealized to a model containing 3 states corresponding to the classical state (C), hybrid state 1 (H1), and hybrid state 2 (H2). (A) tRNAs on the wild-type ribosome are found to occupy predominantly a high-FRET classical configuration with transient excursions to the lower FRET hybrid states. (B) Mutation of residue G2553 of the A loop, and (C) G2252 of the P loop dramatically alter the stability of tRNA configurations on the ribosome, changing the energy landscape to favor intermediate- (B) and low- (C) FRET hybrid states.