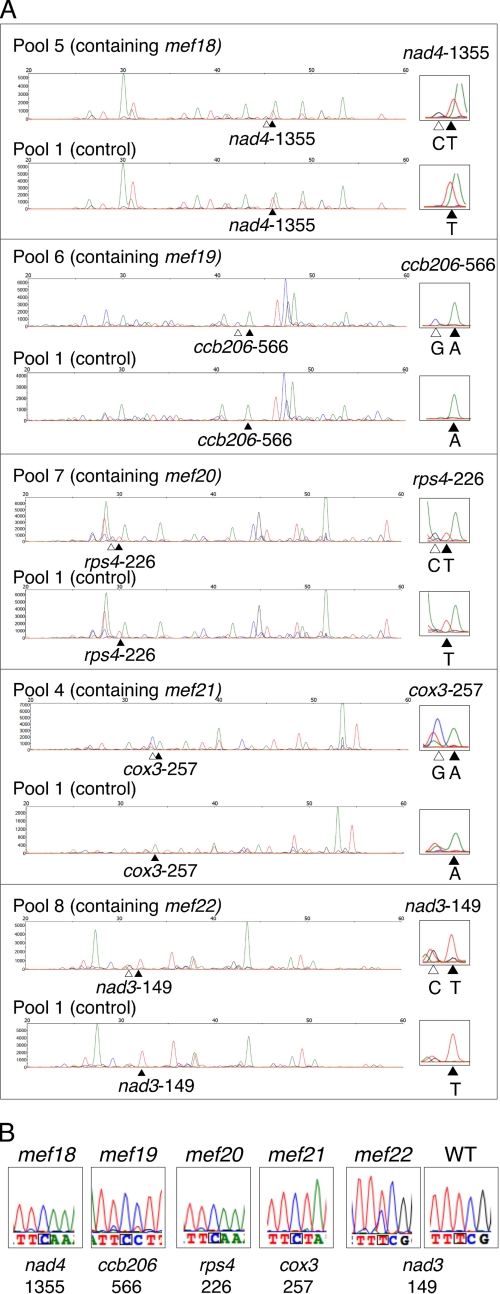
FIGURE 1.
A SNaPshot screen of 58 T-DNA insertion lines of E-class PPR proteins identifies five lines defective in RNA editing at specific sites in mitochondrial RNAs of the mutant plants. A, SNaPshot analyses of RNA-editing sites in the cDNAs of the indicated mitochondrial mRNAs are shown for the identification of the five mutants mef18-mef22 in A. thaliana. The left parts show an analysis of the RNA prepared from pooled leaves of 8 plants each, the top lines contain the mutant line indicated and for comparison of the tracing of pool 1, which contain none of these mutants as a negative control. The relevant sites are shown enlarged on the right hand side with the C and G traces derived from one unedited (or partially edited) plant in the pool indicated by open arrowheads. The filled arrowheads point to the respective T or A traces of the edited nucleotide. The open triangles indicate the respective C or G traces of the nucleotide unedited in the mutant plant. The mutant will be present as one in eight plants and will therefore yield a proportionally lower signal. A sixth T-DNA mutant, MEF8, was identified in parallel by mapping of EMS mutants and will be described elsewhere. B, direct sequence analysis of the disturbed RNA-editing sites in the cDNAs of the five mutants mef18-mef22 shows a complete loss of editing at the sites identified in mutants mef18-mef21, whereas mutant mef22 displays in comparison to a Col wild-type A. thaliana plant (WT) reduced editing of about 40% detectable C to U conversion. The mutant plants were confirmed by PCR to be homozygous for the T-DNA insertion. Color traces are C, blue; T, red; G, black; A, green.