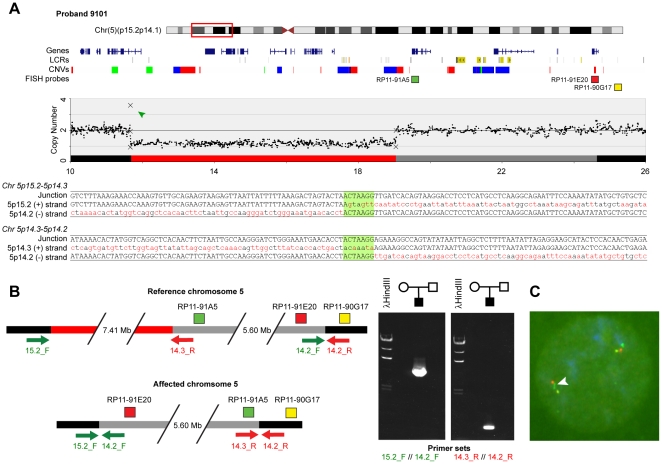
Figure 2. Determination of a complex rearrangement on chromosome 5p15.2-5p14.3 required sequencing deletion breakpoints.
(A) Ideogram of chromosome 5 showing the 5p15.2‐5p14.3 deletion present in proband 9101. The copy number plot was derived from our microarray (black dots) and qPCR (Xs) results. The green arrowhead indicates an inherited CNV at the telomeric deletion breakpoint. Black and gray boxes depict regions of normal copy number, the red box indicates the extent of the deletion, and the gray box shows the inverted region. The sequence tracks show junction fragments along with genomic positions and orientations of flanking sequences. Eight base pairs of 5p14.2 sequence present at both breakpoints are highlighted in green. (B) Schematic of deletion and inversion including relative locations of PCR primers and FISH probes on reference and affected chromosomes 5. Telomeric is to the left. The gel image on the right shows amplification of the junction fragments present at the deletion/inversion breakpoints. The absence from both parents indicates this rearrangement occurred de novo. (C) Metaphase FISH analyses of cells from proband 9101, using BAC probes RP11‐91A5 (green), RP11‐91E20 (red), and RP11‐90G17 (yellow) confirming the presence of the inverted region of chromosome 5p14.5 (white arrowhead).