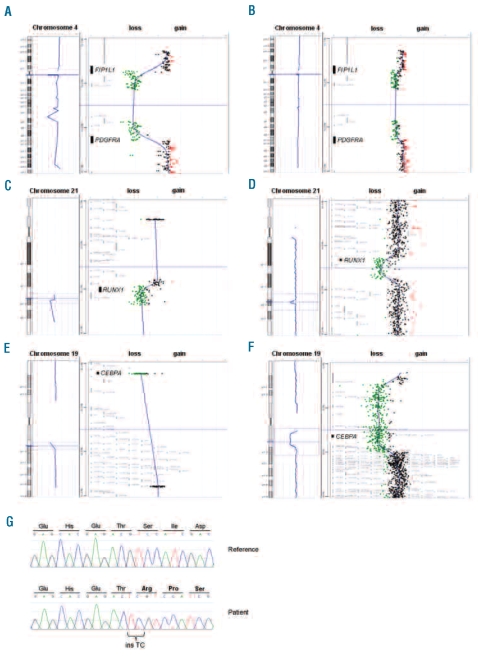
Figure 1.
Targeted array CGH and sequencing profiles. (A) Control experiments demonstrating that targeted arrays were readily able to identify the 800kb FIP1L1-PDGFRA deletion in EOL1 cells in a background of normal cells. EOL1 cells harboring two copies of the del(4) and one normal chromosome 4. (B) 1:2.5 mixtures of EOL1 DNA with normal DNA, simulating a heterozygous deletion with 50% background normal cells. (C) RUNX1 deletion in a patient with atypical CML. Targeted array CGH profile of chromosome 21 (left) with zoom of the respective region (right) showing a hemizygous deletion of RUNX1. (D) Whole-genome 244K array CGH results confirm this observation and further characterize the deletion as a 841kb deletion including RUNX1. (E) CEBPA deletion in a patient with atypical CML. Targeted array CGH profile of chromosome 19 (left) with zoom of the respective region (right) showing a hemizygous deletion of CEBPA. (F) Whole-genome 244K array CGH results further characterize the deletion as a 6.3Mb deletion including CEBPA and several other genes. (G) Sequencing result of the atypical CML patient with a CEBPA deletion. A homozygous 2 bp insertion was observed resulting in a frameshift at amino acid threonine 60 (bottom panel). The CEBPA reference sequence is shown in the top panel.