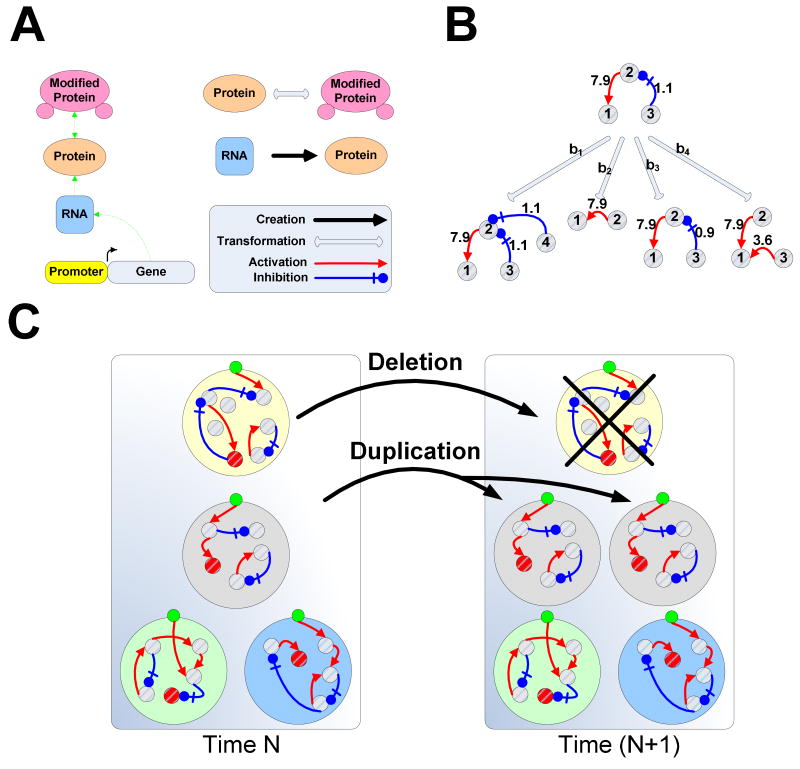
Figure 2. A biochemically realistic simulation framework.
(A) The networks are structured around the ‘central dogma’: activation of gene transcription creates RNA that can be translated to protein, which in turn, can be modified to change its state. (B) Possible mutational scenarios for generic nodes (representing either RNA, protein or modified protein). For example, node 3 can be duplicated (b1), destroyed (b2), undergo mild mutation, changing the strength of its regulation of node 2 (b3), or mutate strongly causing its decoupling from node 2 and coupling to node 1 (b4). In addition to network-topology, mutations may change node parameters such as basal transcription-rate, degradation-rate and interaction affinity. (C) Organisms that accumulate enough energy undergo duplication and increase their representation in the population. Duplication is accompanied by deletion of the organism with the least energy in the population.