FIG. 3.
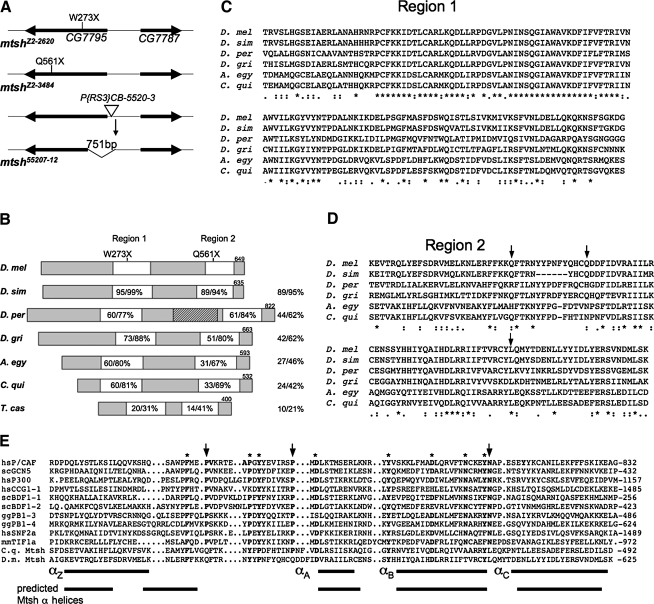
The mitoshell gene corresponds to CG7795 and encodes a protein containing a bromodomain-related region. (A) Schematic depiction of the CG7795 region in polytene interval 29A on chromosome 2, showing the mutations in three mtsh alleles. The Zuker collection alleles mtshZ2-2620 and mtshZ2-3484 each have a unique nonsense mutation in CG7795. Mobilization of the P{RS3}CB-5520-3 homozygous viable and fertile transposable element (triangle) led to the mtsh55207-12 allele that lacks 751 bp, including the transcriptional and translational start sites of CG7795, but not affecting the adjacent gene CG7787. (B) Schematic diagram of Mtsh and its orthologs in 3 of the 11 other sequenced Drosophila species (D. simulans representing the closest neighbor, D. persimilis representing the obscura subgroup, and D. grimshawi representing the most distantly related), as well as Aedes egyptii, Culex quinquefasciatus, and Tribolium castaneum. Protein sizes are indicated at top right of each. The locations of nonsense mutations in mtshZ2-2620 and mtshZ2-3484 are indicated in regions 1 and 2 respectively. Amino acid identity/similarity to D. melanogaster Mtsh is shown at right for each full protein and within the white boxes for the highly conserved regions 1 and 2. The stippled box is a region found only in the obscura group of Drosophila species that resembles a domain of DNA polymerase III subunits tau and gamma. (C) and (D) ClustalW sequence lineups of the highly conserved regions 1 and 2 in Mtsh and Dipteran orthologs. Asterisks indicate identical residues, and double and single dots indicate the strongly and weakly similar residues, respectively. Arrows indicate sites corresponding to those most crucial for binding to acetylated lysine in typical bromodomains; also indicated with arrows in panel (E). (E) Sequence lineup comparing the bromodomain-related region (region 2) in Mtsh and the C. quinquefasciatus ortholog (bottom two rows) with 10 known bromodomains. Adapted from Dhalluin et al. (1999), with bromodomain-containing proteins as described therein. Arrows indicate the most highly conserved bromodomain residues. Asterisks indicate other highly conserved bromodomain residues. At bottom, the known four alpha helical regions of bromodomains are indicated, juxtaposed with the predicted alpha helical regions of Mtsh.