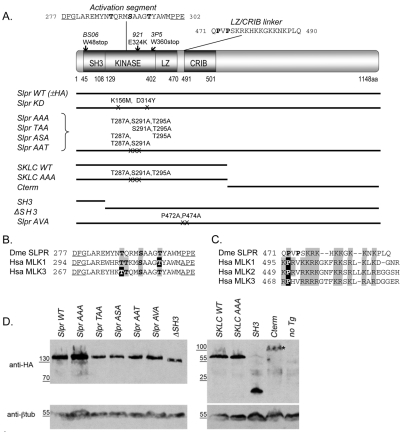
Fig. 1.
Domain architecture and expression of Slpr and derived transgenic constructs. (A) Organization and position of Slpr domains (SH3, Src homology 3; LZ, leucine zipper; CRIB, Cdc42–Rac interactive binding) and mutant alleles. Constructs are shown as black lines with mutated amino acids noted. The kinase activation segment, flanked by conserved residues (underlined), shows putative Ser/Thr phosphorylation targets (bold). The sequence of the LZ–CRIB linker shows the proline residues (bold) that were targeted for mutagenesis. (B) Alignment of the activation segments of Drosophila Slpr (Dme) and its human (Hsa) homologs. Conserved phosphorylation sites are indicated by grey boxes. Black boxes identify experimentally determined primary phosphosites in the human MLKs. (C) Sequence alignment of the LZ–CRIB linkers of Slpr and its human homologs. Note the conserved proline residue(s) followed by a stretch of basic amino acids (grey boxes). (D) Western immunoblots of embryonic lysates expressing HA-epitope-tagged transgenic proteins. β-tubulin was used as a loading control. Transgenic proteins were stably expressed with the exception of the Cterm (asterisk), whose levels are reduced.