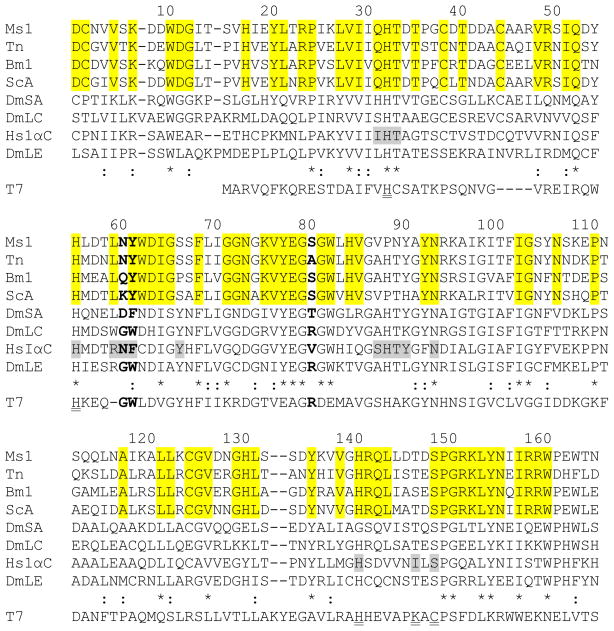
Fig. 1. Multiple sequence alignment of PGRP sequences and T7 lysozyme.
Amino acid sequences of M. sexta PGRP1 (Ms1, AF413068), T. ni PGRP (Tn, AAC31820), B. mori PGRP (Bm1, BAA77209), S. cynthia PGRP-A (ScA, BAF03522), D. melanogaster PGRP-SA (DmSA, NP_572727), -LC (DmLC, NP_729468.2), -LE (DmLE, NP_573078), H. sapiens PGRP-IαC (HsIαC, AAK72484), and T7 lysozyme (T7, NP_0419731) are aligned. Residue numbers of mature M. sexta PGRP1 are indicated on top of its sequence. Identical and similar residues in PGRPs are marked with “*” and “:”, respectively. Residues corresponding to Asn236 and Phe237 of human PGRP-IαC and Arg254 of Drosophila PGRP-LE are shown in bold. The muramyl tripeptide-interacting residues in human PGRP-IαC are shaded gray, residues identical in the lepidopteran PGRPs are highlighted, and residues for Zn2+-binding and amidase activity in T7 lysozyme are double underlined.