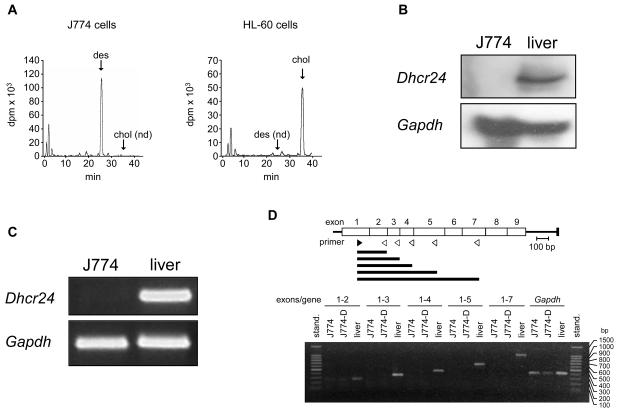
Figure 1.
Sterol biosynthesis and Dhcr24 expression in J774 cells. (A) [14C]Acetate incorporation into sterols in J774 and HL-60 cells. Cells were incubated in RPMI containing 10% LPDS supplemented with 40 μCi of [14C]acetate for 10 h and the cell extracts containing the nonsaponifiable lipid fraction were analyzed by reverse-phase HPLC and in-line radioactivity counting. The arrows indicate the elution times of desmosterol (des), and cholesterol (chol). nd, not detected. (B) Northern blot analysis of RNA of J774 cells and mouse liver. Total RNA was subjected to electrophoresis and blot hybridization with digoxygenin-labelled cDNA probes for mouse Dhcr24 and mouse Gapdh. (C) RT-PCR analysis of RNA of J774 cells and mouse liver for detection of Dhcr24 and Gapdh. (D) RT-PCR analysis of RNA of J774 cells and mouse liver for detection of Dhcr24 cDNA fragments of increasing length and comprising regions expanding from exon 1 up to exon 7, as indicated. J774-D cells are J774 cells grown in DCCM-1 (see text). The upper panel indicates the position of the sense (no. 3 in Supplementary Table S1) and antisense (no. 4-8 in Supplementary Table S1) primers (filled and empty arrowheads, respectively) used for the amplifications and the resulting cDNA fragments (horizontal bars). stand., standard; bp, base pairs.