Abstract
The type III secretion system (T3SS) is an interspecies protein transport machine that plays a major role in interactions of Gram-negative bacteria with animals and plants by delivering bacterial effector proteins into host cells. T3SSs span both membranes of Gram-negative bacteria by forming a structure of connected oligomeric rings termed the needle complex (NC). Here, the localization of subunits in the Salmonella enterica serovar Typhimurium T3SS NC were probed via mass spectrometry-assisted identification of chemical cross-links in intact NC preparations. Cross-links between amino acids near the amino terminus of the outer membrane ring component InvG and the carboxyl terminus of the inner membrane ring component PrgH and between the two inner membrane components PrgH and PrgK allowed for spatial localization of the three ring components within the electron density map structures of NCs. Mutational and biochemical analysis demonstrated that the amino terminus of InvG and the carboxyl terminus of PrgH play a critical role in the assembly and function of the T3SS apparatus. Analysis of an InvG mutant indicates that the structure of the InvG oligomer can affect the switching of the T3SS substrate to translocon and effector components. This study provides insights into how structural organization of needle complex base components promotes T3SS assembly and function.
IMPORTANCE
Many biological macromolecular complexes are composed of symmetrical homomers, which assemble into larger structures. Some complexes, such as secretion systems, span biological membranes, forming hydrophilic domains to move substrates across lipid bilayers. Type III secretion systems (T3SSs) deliver bacterial effector proteins directly to the host cell cytoplasm and allow for critical interactions between many Gram-negative pathogenic bacterial species and their hosts. Progress has been made towards the goal of determining the three-dimensional structure of the secretion apparatus by determination of high-resolution crystal structures of individual protein subunits and low-resolution models of the assembly, using reconstructions of cryoelectron microscopy images. However, a more refined picture of the localization of periplasmic ring structures within the assembly and their interactions has only recently begun to emerge. This work localizes T3SS transmembrane rings and identifies structural elements that affect substrate switching and are essential to the assembly of components that are inserted into host cell membranes.
INTRODUCTION
Gram-negative bacteria have evolved various secretion systems to translocate bacterial effector proteins across the cell envelope to fulfill diverse functions, including some essential for pathogenesis (1). A variety of strategies have been used to obtain structural information about these assemblies, including determination of crystal structures of monomeric soluble domains, three-dimensional reconstructions using cryoelectron microscopy, molecular modeling, and traditional biochemical methods for probing specific interactions (2–11). One of these complex membrane systems is the type III secretion system (T3SS), used by many Gram-negative pathogens to directly deliver bacterial effector proteins to the host cell cytoplasm (12). More than 20 conserved proteins form the T3SS apparatus, with a structural core composed of connected inner membrane (IM) and outer membrane (OM) rings and a protruding extracellular needle termed the needle complex (NC) (2, 3, 13, 14). NCs assemble through the formation of outer and inner membrane rings by sec-mediated secretion. It is unknown how these rings find each other after they form in different membranes. Subsequently, cytoplasmic proteins interact with this ring structure to secrete components that form the secretion channel within the rings, which then serve as the platform for secretion of other components, including the needle, the needle tip, the translocon that is inserted in the eukaryotic membrane, and the effectors, which are ultimately delivered to the eukaryotic cell cytoplasm.
The Salmonella enterica serovar Typhimurium and Shigella flexneri NCs have been purified and examined at approximately 17-Å resolution by cryoelectron microscopy (cryo-EM) with three-dimensional reconstructions, indicating that the assembly consists of two connected membrane ring structures of different diameters (2, 4, 7, 11, 13, 15). The outer membrane (OM) ring has a smaller diameter and is formed by 12 to 14 subunits of a secretin protein family member (InvG, MxiD, and EscC in S. Typhimurium, S. flexneri, and enteropathogenic Escherichia coli [EPEC], respectively) (4, 6, 13, 16). These proteins contain a variable N-terminal periplasmic region and a highly conserved protease-resistant C-terminal region embedded in the OM. The inner membrane (IM) ring has a larger diameter, is composed of a protein from the conserved PrgK/MxiJ/EscJ family, and is likely surrounded by a second protein from the PrgH/MxiG/EscD family (2, 3, 5, 11, 13, 15). In S. Typhimurium, PrgK is anchored to the IM through an N-terminal lipid moiety and a C-terminal transmembrane domain. PrgH, which surrounds PrgK in the IM, has an N-terminal transmembrane domain and a C-terminal periplasmic domain (2, 3, 14). The monomeric crystal structures of the periplasmic domains of the OM secretin EscC as well as the IM proteins EscJ and PrgH have been determined (5, 6). The stoichiometries and symmetries of membrane components are controversial due to the lack of a sufficiently resolved structure showing individual NC subunits. The periplasmic N-terminal domain of the E. coli inner membrane ring EscJ crystallized as a tetrameric asymmetrical unit which oligomerized around the 6-fold helical symmetry axis of the crystal to suggest an intimately associated 24-membered ring, which is consistent with biochemical stoichiometry determinations for S. Typhimurium (5). Recent reports favor an OM-to-IM ring subunit ratio of 1:2, with 12-fold symmetry in the outer membrane rings and 24-fold symmetry for the inner membrane rings in Shigella spp. and S. Typhimurium (5–7, 11), while others reported mass ratios of 1:1 and 20-fold symmetry for S. Typhimurium (4, 15).
Molecular modeling and docking of the EscC and PrgH ring structures within the S. Typhimurium NC cryo-EM maps suggested the possibility that the OM and IM rings could directly interact (6). The N-terminal region of the secretin EscC was shown to span far into the periplasm, though modeling could not distinguish between two potential locations of the secretin in the assembly, given the resolution of the experimental data (6). Alternatively, the OM and IM ring components may simply be linked through the inner rod or needle proteins. This seems unlikely since S. Typhimurium strains defective for the inner rod or needle protein form basal body ring structures lacking needles, suggesting direct interactions between the ring proteins (3).
Here, we purified NCs and performed mass spectrometry (MS)-aided identification of chemically cross-linked and derivatized native T3SS NCs to further resolve ring component interactions (Fig. 1). These results were utilized in conjunction with structure-based modeling to create a model of the organization of periplasmic ring structures in the NC. The model was used to guide functional analyses of critical ring component interactions to gain a better understating of T3SS assembly and regulation.
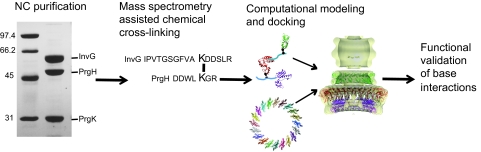
FIG 1 .
Schematic of integrated approach for identifying type III base component interactions. Purified needle complexes were subjected to chemical cross-linking and analyzed by mass spectrometry. Computational modeling and docking made use of base component crystal structures (5, 6), needle complex EM density maps (4, 15), and experimental data. The effects of base component interactions on type III needle complex assembly and function were assessed.
RESULTS
T3SS OM and IM ring components interact.
Purified NCs from S. Typhimurium were chemically cross-linked prior to tryptic digestion and analyzed by a mass spectrometry-based acquisition/analysis pipeline (17). Early attempts to identify cross-links were hampered by significant contamination of NCs with the oligomeric chaperone Hsp60 and ribosomal proteins, as determined by mass spectrometry-based peptide profiling of purified NCs (see Fig. S1 and Tables S1a and S2 in the supplemental material). The addition of a size exclusion chromatography step following cesium chloride density centrifugation allowed for separation of the majority of contaminants from NCs, as evidenced by a reduction in the number of peptides identified and spectral count ratios for NC components (see Fig. S1b and Tables S1b and S2 in the supplemental material).
Chemical cross-linking coupled with mass spectrometry-based identification of NCs with cross-linkers of various spacer arm lengths and reactivities suggested several homotypic and heterotypic interactions between the three ring proteins (Table 1) (17). One homotypic cross-link with succinimidyl-ester diazirine (SDA), a membrane permeable amine to nonselective cross-linker with a 3.9-Å spacer arm, was identified in each of the ring proteins (PrgH Asp251 to PrgH Lys278, PrgK Lys203 to PrgK Lys203, and InvG Lys54 to InvG Lys134). The InvG Lys54-to-InvG Lys134 cross-link was also identified using the water-soluble cross-linker bis[sulfosuccinimidyl] glutarate (BS2G), which has a 7.7-Å spacer arm and uses N-hydroxysuccinimide (NHS) ester reactivity to cross-link amine to amine. The InvG Lys38 residue was also cross-linked to PrgH Tyr387 with SDA, suggesting that the N-terminal domain of InvG and the C-terminal domain of PrgH are in direct contact. This possible interaction was supported by a second cross-link determined using the zero-length cross-linker 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) between InvG Lys38 and the terminal carboxylate of PrgH, Leu392. Four cross-links between the inner membrane components PrgH and PrgK involving residues located in the periplasmic region of PrgH following the transmembrane region and residues of PrgK proximal to the transmembrane domain were identified (Table 1), indicating that the IM components are also likely in intimate contact. These results support interactions between the two IM ring components PrgH and PrgK and, in addition, the idea that the OM ring component InvG and the IM ring component PrgH directly interact.
TABLE 1 .
List of cross-linked peptides identified between NC componentsa
| Exptl mass | Charge state | Theoretical mass | Mass error (ppm) | Peptide sequence 1 | Protein designation 1 | Peptide sequence 2 | Protein designation 2 | Cross-linker |
|---|---|---|---|---|---|---|---|---|
| 2,208.1921 | 4 | 2,208.1897 | 1.11 | ELEVLSQK(278)LR | PRGH | IHFD(251)E(252)PR | PRGH | SDA |
| 2,469.408 | 4, 5 | 2,469.4104 | 0.99 | KELEVLSQK(278)LR | PRGH | K(255)PVFWLSR | PRGH | BS2G |
| 3,015.5791 | 4 | 3,015.6096 | 10.12 | SDAQLQAPGT(200)PVKR | PRGK | SDAQLQAPGTPVK(203)R | PRGK | SDA |
| 3,015.5791 | 4 | 3,015.6096 | 10.12 | SDAQLQAPGTPVK(203)R | PRGK | SDAQLQAPGTPVK(203)R | PRGK | SDA |
| 3,029.5B59 | 4 | 3,029.5889 | 0.97 | SDAQLQAPGTPVK(203)R | PRGK | SDAQLQAPGTPVK(203)R | PRGK | BS2G |
| 2,076.1345 | 4 | 2,076.1375 | 1.06 | E(55)PVIVSK | INVG | SGLYNK(134)NYPLR | INVG | EDC |
| 3,455.8123 | 4 | 3,455.8159 | 1.06 | TFFDAMALQLK(54)EPVIVSK | INVG | SGLYNK(134)NYPLR | INVG | BS2G |
| 3,160.574 | 4 | 3,160.5688 | 1.62 | IPVTGSGFVAK(38)DDSLR | INVG | MSPGHWY(387)FPSPL | PRGH | SDA |
| 3,060.5195 | 4 | 3,059.5835 | 5.45 | IPVTGSGFVAK(38)JDOSLR | INVG | MSPGHWYFPSPL(392) | PRGH | EDC |
| 2,645.3428 | 4 | 2,645.3445 | 0.65 | IPVTGSGFVAK(38)DDSLR | INVG | DDWLK(367)GR | PRGH | BS2G |
| 2,949.542 | 4 | 2,949.54 | 0.66 | QAA(171)ELDSLLGQEK | PRGH | SDAQLQAPGTPVK(203)R | PRGK | SDA |
| 2,849.4839 | 4 | 2,849.4888 | 1.71 | QAAE(172)LDSLLGQEK | PRGH | SDAQLQAPGTPVK(203)R | PRGK | EDC |
| 2,949.5432 | 4 | 2,949.54 | 1.08 | QAAELDSL(176)LGQEK | PRGH | SDAQLQAPGTPVK(203)R | PRGK | SDA |
| 3,134.6287 | 4 | 3,134.6326 | 1.25 | QAAELDSLLGQE(180)KER | PRGH | SDAQLQAPGTPVK(203)R | PRGK | EDC |
| 3,248.6577 | 4 | 3,248.6631 | 1.65 | QAAELDSLLGQEK(181)ER | PRGH | SDAQLQAPGTPVK(203)R | PRGK | BS2G |
| 2,486.2532 | 4 | 2,486.251 | 0.68 | G(214)DYDKNAR | PRGH | SDAQLQAPGTPVK(203)R | PRGK | SDA |
| 2,336.1982 | 4 | 2,386.1995 | 0.51 | GD(215)YDKNAR | PRGH | SDAQLQAPGTPVK(203)R | PRGK | EDC |
| 2,500.2236 | 4 | 2,500.23 | 2.54 | GDYDK(218)NAR | PRGH | SDAQLQAPGTPVK(203)R | PRGK | BS2G |
The position of each cross-linked residue is indicated in parentheses.
Ring protein interactions are necessary for NC assembly and T3 secretion.
To understand the importance of the identified cross-links in the context of published T3SS NC structural information, we modeled ring structures symmetrically, using the partial structures of PrgH, EscJ (EPEC PrgK homolog), and EscC (EPEC InvG homolog) with the macromolecular modeling suite Rosetta (5, 6, 8). For each ring system, several structural ensembles corresponding to different crystal forms and oligomerization states were generated (12 to 14 for EscC, 20 for EscJ, and 24 for PrgH). A subset of models with both favorable all-atom energy and significant numbers of atomic contacts between protein subunits were fitted into the electron density maps corresponding to isolated rings from the Salmonella and Shigella cryo-EM models. To create the final model (Fig. 2a and b), the conformation and localization of each ring system in the assembly were taken from the molecular model with the highest correlation between modeled and experimental electron density. Ring models of InvG, PrgH, and PrgK were generated from these models by use of homology modeling (6). Stoichiometries of 20 subunits of PrgH and PrgK in the Salmonella cryo-EM model and 24 subunits of PrgH and PrgK in the Shigella model were found to best explain the experimental electron density. For InvG, it was necessary to use a ring with 14 subunits fit into the density map closest to the inner membrane. With only 12 subunits, all ring models are considerably smaller than what is observed for experimental density. If the true stoichiometry is 14, this suggests that additional protein residues, not in the crystallized portion of EscC, contribute to the density in this region. A number of cross-links were useful in validating the molecular models of the ring structures. In particular, cross-links between/within PrgH subunits could be interpreted with the help of the PrgH ring model (see Fig. S3 in the supplemental material). Many of the chemical cross-links to the inner membrane proteins PrgH and PrgK occur outside the regions crystallized; specifically, the PrgH170-362 structure lacks the C-terminal 30 amino acids (aa) while the EscJ21-190 structure lacks the region proximal to the membrane. However, the projected locations of the terminal segments, based on the placement of the N and C termini of PrgK and PrgH, are consistent with the distances observed from the cross-linking experiments. In addition, the locations of the termini are consistent with anchoring of the transmembrane helices of PrgK and PrgH in the membrane.
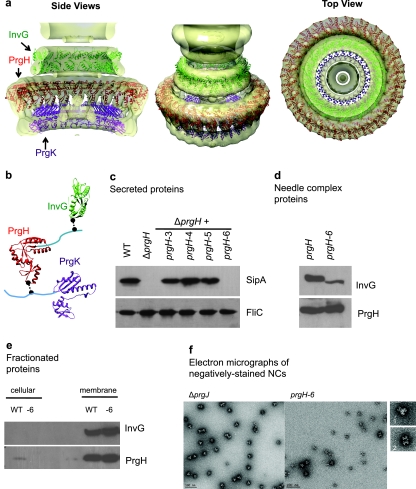
FIG 2 .
T3SS base component interactions. (a) Ring models of InvG (green), PrgH (red), and PrgK (purple) periplasmic domains were docked into the EM density maps of the S. Typhimurium needle complex (4, 15). Models were based on crystal structures of EscC (E. coli homolog of InvG), PrgH (6), and EscJ (E. coli homolog of PrgK) (5) and evaluated against experimentally derived chemical cross-linking and limited biotinylation data. A representative model of the final subset of validated models fulfilling modeling, experimental, and docking constraints is shown. (b) Ring component subunits and proposed docking positions highlighting cross-linked residues (black spheres) between components in regions of unknown structure (cyan). PrgH (residues 363 to 392) and PrgK (residues 191 to 392) fall outside the crystal structures. (c) Effect of PrgH C-terminal deletions on type III secretion by SipA secretion profiles. Secreted proteins from prgH-deficient strains complemented with plasmid encoding prgH C-terminal deletions were analyzed by Western blotting using a monoclonal antibody against SipA and an antibody against the flagellin protein FliC to control for loading. (d) Effect of PrgH C-terminal deletion on type III needle complex assembly. Wild-type and PrgH-6 needle complexes were isolated and analyzed by Western blotting using antibodies against InvG and PrgH. (e) Effect of PrgH C-terminal deletion on type III ring components. Cellular and membrane fractions isolated from S. Typhimurium were analyzed by Western blotting using antibodies against InvG and PrgH. (f) Electron micrographs of negatively stained needle complexes from ΔprgJ and PrgH-6 S. Typhimurium strains with representative structures, showing side views (panel 1, ΔprgJ, and panel 2, PrgH-6).
These results suggested that the C terminus of PrgH and the N terminus of InvG may interact to promote polymerization and assembly of the two rings or to maintain their assembly before needle protein secretion and polymerization. This idea was directly tested by analysis of the C terminus of PrgH, since 2 of the last 6 aa residues in PrgH were cross-linked to InvG. Substitution of alanine for all identified cross-linked residues, including PrgH Tyr387 and PrgH Leu392, did not alter secreted-protein profiles (data not shown). Therefore, strains with sequential amino acid deletions from the C-terminal end of PrgH were generated and assessed for NC assembly and secretion (Fig. 2c). The use of Salmonella strains expressing constructs with deletions of up to five residues from the C terminus of PrgH had little to no effect, while deletion of six residues (PrgH-6) or more, starting with Tyr387, which was cross-linked to InvG Lys38, abolished T3 secretion (Fig. 2c). Analysis of NCs purified from a Salmonella strain expressing PrgH lacking its last 6 aa showed that NC formation is also severely inhibited (Fig. 2d). To determine if abolished secretion and reduced NC formation are due to depleted membrane components or changes in membrane component localization, we assessed the cellular and membrane levels of InvG and PrgH in this mutant and determined that the protein levels and localization are similar to those observed for a Salmonella strain expressing wild-type PrgH (Fig. 2e). Electron micrographs of negatively stained NCs prepared from this mutant strain showed oligomeric structures that may be found to correspond to the IM rings when compared to the structures observed in NCs prepared from Salmonella ΔprgJ, which produces NC basal bodies that lack needles, simplifying comparison of base structures (Fig. 2f). PrgH/PrgK complexes from recombinant expression in E. coli (3) have also been isolated, indicating that the IM ring proteins can oligomerize in the absence of the OM ring protein InvG and that the defect in NC assembly in the PrgH-6 mutant is likely due to a lack of interaction between PrgH and InvG. These results indicate that the last six residues of PrgH are necessary for NC assembly and T3SS function, most likely through direct interaction with the N-terminal domain of the OM component InvG to allow assembly of a competent secretion complex.
The InvG oligomer structure affects substrate switching.
To further understand the role of InvG in NC assembly, we tested surface lysine accessibility. Purified NCs were derivatized with a lysine-specific biotin reagent, sulfo-NHS-LC-biotin, prior to preparation and analysis by mass spectrometry. Derivatization of NC preparations indicated that the very N-terminal domain of the secretin InvG (Lys38, Lys54, and Lys61) was protected from biotinylation, being labeled only in denatured NCs, while lysine residues further from the N terminus (Lys127, Lys 134, and Lys201) were accessible, being labeled both in native and in denatured NCs (Table 2). InvG oligomeric complexes, also purified and treated, showed that lysine residues protected from modification in the native oligomer (Lys61, Lys67, Lys134, and Lys259) extend from the N terminus to the border of the C-terminal protease resistance domain defined previously as Lys269 in NC (6). These results demonstrate that the surface accessibility of InvG differs in NCs, compared to the oligomeric assembly of InvG in the absence of the inner membrane ring, and support the idea that the N-terminal region of InvG is involved in either homo- or hetero-oligomeric interactions within the NC.
TABLE 2 .
List of biotinylated peptides identified in InvG within the needle complexa
| Exptl mass | Charge state | Peptides identified | Denatured NC | Native NC | Labeled residue(s) |
|---|---|---|---|---|---|
| 645.336 | 3 | NVSLNEFNNFLKR | + | + | Lys-127 |
| 702.69 | 3 | SGLYNKNYPLR | + | + | Lys-134 |
| 847.091 | 3 | QSGAWSGDDKLQKWVR | + | + | Lys-545, Lys-548 |
| 1,020.498 | 3 | KGTFYVSGPPVYVDMWNAATMMDK | + | + | Lys-144, Lys-168 |
| 661.027 | 3 | DQKMVIPGIATAIER | + | Lys-201 | |
| 778.42 | 2 | EPVIVSKMAAR | + | Lys-61 | |
| 492.762 | 2 | GQEAIK | + | Lys-562 | |
| 815.937 | 2 | VYLDRGQEAIK | + | Lys-562 | |
| 1,001.027 | 2 | IPVTGSGFVAKDDSLR | + | Lys-38 | |
| 1,048.889 | 3 | TFFDAMALQLK EPVIVSKMAAR | + | Lys-54, Lys-61 | |
| 783.921 | 2 | TFYTKLIGER | + | Lys-392 | |
| 585.827 | 2 | QKIGVMR | + | Lys-179 | |
| 729.723 | 3 | QKIGVMRLNNTFVGDR | + | Lys-179 |
Purified NCs were treated with a lysine specific biotin reagent directly (native NC) or after denaturation in SDS (denatured NC) prior to preparation for LTQ-Orbitrap mass spectrometry. The peptides identified were ±5 ppm in terms of mass accuracy.
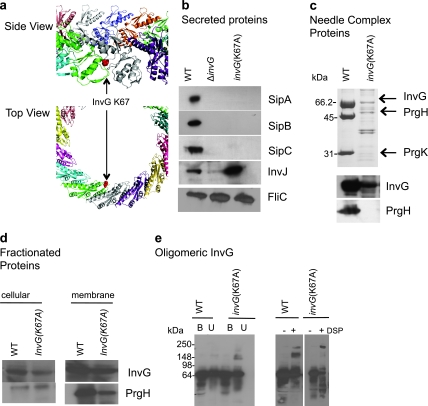
Our validated model also predicted that in addition to the InvG N-terminal loop with Lys38 that was shown to be in close contact with the C terminus of PrgH, a second N-terminal loop (residues 61 to 68) may play an important role in NC assembly or function on the basis of its orientation and proximity to the other NC components (Fig. 3a). We performed alanine-scanning mutagenesis on this loop by complementing an S. Typhimurium ΔinvG strain with InvG loop point mutations and analyzing secretion profiles by Western blotting against SipA (see Fig. S2 in the supplemental material). The results showed that strains carrying InvG(Lys67Ala) do not secrete SipA, while all other point mutants secrete SipA in amounts similar to those observed for the wild type. To confirm this result, secretion profiles from an S. Typhimurium strain with an InvG(Lys67Ala) mutation on the chromosome were analyzed by Western blotting. S. Typhimurium InvG(Lys67Ala) had an unusual secretion profile; secretion of the needle length regulator protein InvJ is increased while secretion of the effector and translocon components SipA, SipC, and SipC was abolished (Fig. 3b). S. Typhimurium ΔinvJ strains produce NCs with abnormally long needles (15). Completion of the inner rod requires InvJ, and in the absence of the protein, it is thought that the inner rod fails to assemble and consequently that the secretion machine is locked in a secretion mode competent for secretion of the needle protein, resulting in needles of altered length. NCs purified from the InvG(Lys67Ala) mutant were isolated in very small quantities, as shown by Western blotting against InvG and PrgH (Fig. 3c), and in insufficient amounts for negative staining and imaging by electron microscopy. Despite this limitation, these results indicate that NC assembly or the structural stability of the complex is affected. The NC components InvG and PrgH were shown to reach the membrane, although the amounts of PrgH were reduced (Fig. 3d). The stability of the InvG(Lys67Ala) oligomer was tested by isolating outer membrane fractions of E. coli C41 cells expressing wild-type InvG or InvG(Lys67Ala) and subjecting samples to boiling as well as chemical cross-linking with dithiobis[succinimidyl propionate] (DSP) prior to Western blotting against the InvG protein (Fig. 3e). The results show that InvG(Lys67Ala) has more-prominent, higher-molecular-weight bands in both unboiled and cross-linked samples, indicating that this mutant may either stabilize the InvG oligomer or increase its oligomerization. We speculate that this may alter InvG interactions with IM rings to stabilize the InvG oligomer and lessen its interactions with the IM rings. Cumulatively, these results indicate that not only are the N-terminal domain of InvG and the C terminus of PrgH in intimate contact but both play critical roles in NC assembly and secretion.
FIG 3 .
InvG affects substrate switching. (a) InvG ring model, with each InvG monomer shown in a different color, with the Lys67 residue highlighted in red. (b) Effect of InvG(Lys67Ala) on type III secretion profiles. Secreted proteins for the invG(Lys67Ala) strain were Western blotted with monoclonal antibody to the effector SipA and transclocon components SipB and SipC and polyclonal antibody against the needle length regulator protein InvJ. Secreted proteins were analyzed by Western blotting using an antibody against the flagellin protein FliC to control for loading. (c) Effect of InvG(Lys67Ala) on type III needle complex assembly. Wild-type and mutant needle complexes were isolated and separated by SDS-PAGE and Western blotted with antibodies to InvG and PrgH. (d) Effect of InvG(Lys67Ala) on type III ring components. Cellular and membrane fractions isolated from S. Typhimurium were analyzed by Western blotting with antibodies against InvG and PrgH. (e) Effect of InvG(Lys67Ala) on the assembly of the InvG oligomer. Outer membrane fractions from E. coli C41 cells expressing InvH and wild-type InvG or InvG(Lys67Ala) were isolated. Samples were boiled in sample buffer (B) or subjected directly (U) to SDS-PAGE and Western blotting with antibody against InvG. Samples were also incubated with or without the cross-linker DSP (dithiobis[succinimidyl propionate]) for 60 min at 22°C prior to SDS-PAGE and Western blotting with antibody against InvG.
DISCUSSION
The spatial localization and orientation of needle complex proteins have begun to be deciphered. Detailed placement of soluble domain structures of ring components into available electron density maps of the needle complex has been hampered by the intermediate resolution of these maps. Recently, insights into the topology and organization of ring components have been provided using a multipronged approach to unambiguously show that InvG forms the outer ring and reaches deep into the periplasmic space to make direct contacts with the inner membrane protein PrgH (11), as has been suggested previously (6). The locations of the N and C termini of PrgH and the N terminus of PrgK, by use of polyhistidine-tagged proteins combined with nanogold labeling and cryoelectron microscopy, have now been clearly shown (11). We have independently confirmed the suggested terminal locations for PrgH and PrgK, with cross-links between two IM components clustering close to the transmembrane regions. While atomic structural models of InvG and PrgK domains have been generated based on homology modeling and ring models of EscJ and EscC have been assembled and docked (5–8, 11), no rigorous modeling of the entire NC base has been attempted and validated. We have constructed a validated NC model using chemical cross-linking and available structures of ring component domains coupled with rigorous computational molecular modeling and docking that further defines the organization and interactions of ring components within the NC.
Both PrgH/PrgK complexes and the InvG oligomer have been isolated in small quantities in the absence of the other membrane ring (3, 6), indicating that oligomerization can occur independently of NC assembly. However, the small amounts of oligomeric membrane rings isolated may indicate that the stability of these rings requires interactions between the IM and the OM during NC base assembly. Our results predict a role for the N-terminal domain of InvG and the C-terminal domain of PrgH in the assembly of the T3SS apparatus and favor an assembly process that includes the secretin binding the inner membrane ring components as an early intermediate and possibly promoting polymerization and or stabilization of both rings, since it is likely disadvantageous to form either ring in the absence of the other. Our results are the first to experimentally demonstrate interactions between the N-terminal domain of InvG and the very C-terminal residues of PrgH. We have clearly defined the requirement of the C-terminal PrgH for assembly of the IM rings, with the OM ring necessary for competent secretion. This result also suggested that the InvG N-terminal loops that are oriented down in the periplasm towards the IM may play a role in assembly or function. In addition to the InvG Lys38 residue interacting with the C terminus of PrgH, we demonstrated that the InvG(Lys67Ala) mutation increases secretion of the needle length regulator InvJ and did not promote secretion of translocon and effector proteins. The NC formation of this mutant was also altered, and the stability of the purified InvG oligomer may be increased. This may suggest that the InvG oligomer structure plays a role within the NC in substrate switching and that different NC conformational states exist for secretion of the needle versus subsequent translocon and effector proteins. This suggestion is intriguing in light of recent results in the SPI2 T3SS where the association state of a cytoplasmic regulatory complex acts to select secretion substrates in response to pH (18). While the identity of the pH sensor is not known, the SPI-2 T3SS is suggested to undergo conformational change on exposure to neutral pH to transduce a dissociation signal to the cytoplasmic regulatory complex to allow effector protein secretion (18). The current study, coupled with a recent study conducted by the Marlovits group (11), builds on available structural information using integrated experimental approaches to provide a structural framework for more-detailed structural studies that will gradually reveal the molecular structure of the type III secretion system apparatus.
MATERIALS AND METHODS
Bacterial strains, construction, and growth conditions.
S. Typhimurium strains ATCC 14028s and TK385, a needle complex-overexpressing strain, were used for secreted-protein analysis and NC purification, respectively (3). Site-directed mutagenesis was carried out using a QuikChange mutagenesis kit (Stratagene). S. Typhimurium prgH deletion strains TK164 and TK359 (3) were complemented with pTK52 (pWSK29 carrying prgH) (3) or pWSK29prgH-6 for secreted protein analysis or NC purification, respectively. Salmonella strains were grown in LB medium and LB supplemented with 300 mM NaCl for needle complex purification. Bacterial strains were constructed using the λ-RED system (19).
Needle complex expression and purification.
NCs were prepared as described previously (15), with the addition of gel filtration on a Superose 6 (GE Healthcare) column in 1× phosphate-buffered saline (PBS), 0.5 M NaCl, 0.2% LDAO (N,N-Dimethyldodecylamine-N-oxide) as a final purification step.
Needle complex cross-linking.
Purified NC was treated with various cross-linkers under the following conditions. NC (50 µg) was treated with 1 mM bis[sulfosuccinimidyl]glutarate (BS2G) diluted from a 100 mM stock solution in dimethyl sulfoxide (DMSO) for 2 h at room temperature, followed by addition of 50 mM Tris-HCl, pH 8.8, to quench the reaction. NC was treated with 1 mM NHS-LC-diazirine or NHS diazirine for 2 h on ice, followed by dialysis against the Superose 6 buffer overnight, to remove unreacted cross-linker. NC was treated with 5 mM 1-ethyl-3-[dimethylaminopropyl]carbodiimide HCl (EDC) and 5 mM sulfo NHS overnight at room temperature, followed by addition of 50 mM Tris-HCl, pH 8.8, to quench the reaction. All samples were denatured with 6 M urea, treated with 0.5 mM TCEP [Tris(2-carboxyethyl)phosphine Hydrochloride] to reduce disulfide bonds, followed by treatment with 40 mM iodoacetamide to prevent disulfides from reforming. All samples were dialyzed extensively against 50 mM ammonium bicarbonate to remove detergent, salt, urea, and other components that would interfere with MS analysis. Samples were digested with substrate-trypsin at a 30:1 (wt/wt) ratio overnight at 37°C, followed by MS analysis.
Cross-linked needle complex mass spectrometry.
Peptide digests were analyzed with a hybrid linear ion trap-Orbitrap instrument (Thermo, Fisher, San Jose, CA) using the chemical cross-linking and mass spectrometry (CXMS) pipeline described elsewhere (17). Briefly, data-dependent ion selection was triggered on peptides with charge states of ≥4+, followed by tandem MS acquisition with the Orbitrap analyzer. Tandem mass spectra were deconvoluted and searched with Phenyx (20) to identify and remove the spectra matching non-cross-linked, linear peptides. Remaining spectra were searched using Popitam (21) in an “open-search mode” to identify cross-linked peptides.
Computational methodology.
Models of the oligomeric forms of PrgH, PrgK, and InvG were produced using the symmetrical assembly protocols (8) in the Rosetta macromolecular modeling suite. The protocol takes as input the three-dimensional structure of the single subunit and optimizes the total energy of the protein complex while varying the degrees of freedom of the rigid body, side chain, and limited backbone (in the case of EscC/InvG). The ring structures were assumed to be perfectly symmetrical, and different crystal forms and oligomerization states were tested in independent simulations (12 to 14 for EscC and 20 and 24 for PrgK and PrgH). The modeling of PrgH, PrgK, and InvG was performed using the crystal structures of the EPEC proteins, followed by homology modeling to create the Salmonella variants. For InvG (EscC), the conformation of the linker between the N- and C-terminal domains of the crystallized portion was also varied as described in reference 6. The results of the simulations for PrgK (EscJ) are found in reference 8. Out of the 5,000 to 30,000 models generated in each simulation, a smaller subset, having low energy and significant numbers of contacts between subunits, was selected for screening by electron density fitting (250 to 1,000 models per simulation). For the density fitting, individual rings were manually isolated from the Salmonella and Shigella electron density maps by use of Chimera (22). For EscC, only the density map closest to the inner membrane was extracted. The oligomer models were ranked based on the cross-correlation score between the predicted density from the model and the density from the experimental data by use of a high-throughput fitting method (23). The best-fitting models by this measure were selected for the final prediction. For EscJ/PrgK, a more confident model of its ring form exists (8), which was fitted into the density map below InvG by use of Situs (24).
Secretion assays and needle complex component analysis.
Secreted proteins were prepared as described elsewhere (3). Purified NCs and cellular fractions were run on a 10% SDS-PAGE gel and subjected to Western blotting as described previously (25).
Electron microscopy and image processing.
To image samples by electron microscopy, purified NCs were negatively stained with 0.75% uranyl formate as described previously (26). All samples were imaged with a transmission electron microscope (Morgagni M268; FEI, Hillsboro, OR) equipped with a tungsten filament and operating at an acceleration voltage of 100 kV. All images were recorded on a 4,000- by 2,000-pixel UltraScan charge-coupled-device (CCD) camera (Gatan, Inc.) at a magnification of ×22,000.
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants U54 AI05741 and R01 AI030479 (to S.I.M.), P20 GM076222 (to D.B.), and 1U54 AI57141-01 (to D.R.G.) and by National Cancer Institute grant R33CA099139-01 (to D.R.G.). S.S. is supported by the Natural Sciences and Engineering Research Council of Canada and I.A. by the Knut and Alice Wallenberg Foundation. T.G. is a Howard Hughes Medical Institute Early Career scientist. N.C.J.S. Is a Howard Hughes International Scholar and thanks the Canadian Institute of Health Research for funding.
We thank E. E. Galyov (University of Leicester) for providing the SipA antibody, V. Koronakis (University of Cambridge) for providing the InvG antibody, the Murdock Charitable Trust and the Washington Research Foundation for generous support of our electron cryomicroscopy facility, and Calvin Yip for helpful discussions.
Footnotes
Citation Sanowar, S., P. Singh, R. A. Pfuetzner, I. André, H. Zheng, et al. 2010. Interactions of the transmembrane polymeric rings of the Salmonella enterica serovar Typhimurium type III secretion system. mBio 1(3):e00158-10. doi:10.1128/mBio.00158-10.
REFERENCES
- 1. Saier M. H., Rodgers L., Tamang D. G., Yen M. R. 2008. Protein secretion and membrane insertion systems in bacteria and eukaryotic organelles. Adv. Appl. Microbiol. 65:141–197 [DOI] [PubMed] [Google Scholar]
- 2. Kubori T., Matsushima Y., Nakamura D., Uralil J., Lara-Tejero M., Sukhan A., Galan J. E., Aizawa S. I. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605 [DOI] [PubMed] [Google Scholar]
- 3. Kimbrough T. G., Miller S. I. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U. S. A. 97:11008–11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marlovits T. C., Kubori T., Sukhan A., Thomas D. R., Galan J. E., Unger V. M. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306:1040–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yip C. K., Kimbrough T. G., Felise H. B., Vuckovic M., Thomas N. A., Pfuetzner R. A., Frey E. A., Finlay B. B., Miller S. I., Strynadka N. C. 2005. Structural characterization of the molecular platform for type III secretion system assembly. Nature 435:702–707 [DOI] [PubMed] [Google Scholar]
- 6. Spreter T., Yip C. K., Sanowar S., Andre I., Kimbrough T. G., Vuckovic M., Pfuetzner R. A., Deng W., Yu A. C., Finlay B. B., Baker D., Miller S. I., Strynadka N. C. 2009. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16:468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hodgkinson J. L., Horsley A., Stabat D., Simon M., Johnson S., da Fonseca P. C., Morris E. P., Wall J. S., Lea S. M., Blocker A. J. 2009. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat. Struct. Mol. Biol. 16:477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andre I., Bradley P., Wang C., Baker D. 2007. Prediction of the structure of symmetrical protein assemblies. Proc. Natl. Acad. Sci. U. S. A. 104:17656–17661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Korotkov K. V., Pardon E., Steyaert J., Hol W. G. 2009. Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17:255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chandran V., Fronzes R., Duquerroy S., Cronin N., Navaza J., Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schraidt O., Lefebre M. D., Brunner M. J., Schmied W. H., Schmidt A., Radics J., Mechtler K., Galan J. E., Marlovits T. C. 2010. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 6:e1000824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blocker A., Jouihri N., Larquet E., Gounon P., Ebel F., Parsot C., Sansonetti P., Allaoui A. 2001. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39:652–663 [DOI] [PubMed] [Google Scholar]
- 14. Kimbrough T. G., Miller S. I. 2002. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 4:75–82 [DOI] [PubMed] [Google Scholar]
- 15. Marlovits T. C., Kubori T., Lara-Tejero M., Thomas D., Unger V. M., Galan J. E. 2006. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441:637–640 [DOI] [PubMed] [Google Scholar]
- 16. Genin S., Boucher C. A. 1994. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet. 243:112–118 [DOI] [PubMed] [Google Scholar]
- 17. Singh P., Shaffer S. A., Scherl A., Holman C., Pfuetzner R. A., Larson Freeman T. J., Miller S. I., Hernandez P., Appel R. D., Goodlett D. R. 2008. Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal. Chem. 80:8799–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu X. J., McGourty K., Liu M., Unsworth K. E., Holden D. W. 2010. pH sensing by intracellular Salmonella induces effector translocation. Science 328:1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Colinge J., Masselot A., Giron M., Dessingy T., Magnin J. 2003. OLAV: towards high-throughput tandem mass spectrometry data identification. Proteomics 3:1454–1463 [DOI] [PubMed] [Google Scholar]
- 21. Hernandez P., Gras R., Frey J., Appel R. D. 2003. Popitam: towards new heuristic strategies to improve protein identification from tandem mass spectrometry data. Proteomics 3:870–878 [DOI] [PubMed] [Google Scholar]
- 22. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 23. DiMaio F., Soni A., Phillips G., Shavlik J. 2007. Improved methods for template-matching in electron-density maps using spherical harmonics. Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine, Fremont, CA.
- 24. Wriggers W. 2010. Using Situs for the integration of multi-resolution structures. Biophys. Rev. 2:21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pegues D. A., Hantman M. J., Behlau I., Miller S. I. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169–181 [DOI] [PubMed] [Google Scholar]
- 26. Ohi M., Li Y., Cheng Y., Walz T. 2004. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced. Online 6:23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.