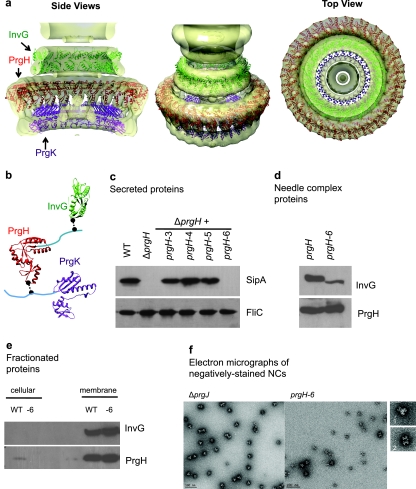
FIG 2 .
T3SS base component interactions. (a) Ring models of InvG (green), PrgH (red), and PrgK (purple) periplasmic domains were docked into the EM density maps of the S. Typhimurium needle complex (4, 15). Models were based on crystal structures of EscC (E. coli homolog of InvG), PrgH (6), and EscJ (E. coli homolog of PrgK) (5) and evaluated against experimentally derived chemical cross-linking and limited biotinylation data. A representative model of the final subset of validated models fulfilling modeling, experimental, and docking constraints is shown. (b) Ring component subunits and proposed docking positions highlighting cross-linked residues (black spheres) between components in regions of unknown structure (cyan). PrgH (residues 363 to 392) and PrgK (residues 191 to 392) fall outside the crystal structures. (c) Effect of PrgH C-terminal deletions on type III secretion by SipA secretion profiles. Secreted proteins from prgH-deficient strains complemented with plasmid encoding prgH C-terminal deletions were analyzed by Western blotting using a monoclonal antibody against SipA and an antibody against the flagellin protein FliC to control for loading. (d) Effect of PrgH C-terminal deletion on type III needle complex assembly. Wild-type and PrgH-6 needle complexes were isolated and analyzed by Western blotting using antibodies against InvG and PrgH. (e) Effect of PrgH C-terminal deletion on type III ring components. Cellular and membrane fractions isolated from S. Typhimurium were analyzed by Western blotting using antibodies against InvG and PrgH. (f) Electron micrographs of negatively stained needle complexes from ΔprgJ and PrgH-6 S. Typhimurium strains with representative structures, showing side views (panel 1, ΔprgJ, and panel 2, PrgH-6).