FIG. 2.
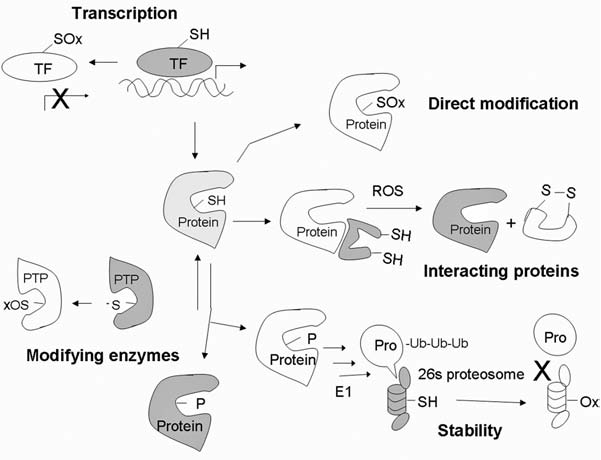
Redox-mediated mechanisms that regulate protein functions. Protein expression can be regulated through redox modification of transcription factors. Oxidation of Cys at or near the DNA-binding site may disrupt the transactivation activity. Newly synthesized protein can be directly modified by oxidation of amino acids such as Cys, Tyr, and Met, resulting in alteration of the protein functions. Certain proteins are stabilized by their redox-sensitive interacting proteins. Modification of the interacting proteins can dissociate the complex and allow activation of the functional proteins. Posttranslational modifications such as phosphorylation can either activate or inhibit protein functions. Phosphatases, which are responsible for dephosphorylation, can be oxidatively inactivated, promoting phosphorylation of proteins. Stability of signaling proteins determines both the level and duration of their functional effects. Most proteins can be degraded through the ubiquitin–proteosome system. Ubiquitin-activating enzyme E1 and proteosome 26S and 20S can be inactivated under oxidative stress. TF, transcription factor; -SH, reduced thiol; SOx, oxidized thiol; PTP, protein tyrosine phosphatase; ub, ubiquitin. X, inhibition; white, inactive state; light grey, partially activated; dark grey, fully activated molecules.
