Figure 1.
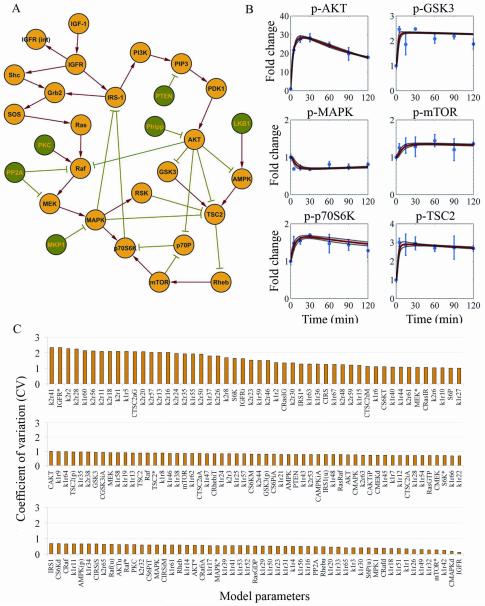
(A) IGFR signaling network topology in the MDA-MB231 cell line. Nodes represent proteins, edges represent protein interactions, red arrows describe protein activation, and green plungers describe protein inactivation. (B) Protein profiles were measured on RPPA in triplicate. The mean protein profiles of non-IGF-1–stimulated cells were used as controls for normalization. Circles and error bars represent the mean and SD of the normalized protein profiles. Normalized time courses were computationally evaluated using the trained mass-action model. Solid red lines represent the mean time courses of the trajectories that equally fit the experimental data, and dashed black lines represent the fitting variability. (C) Histogram of model parameter regimens clustered according to the coefficient of variation (CV=SD/mean).