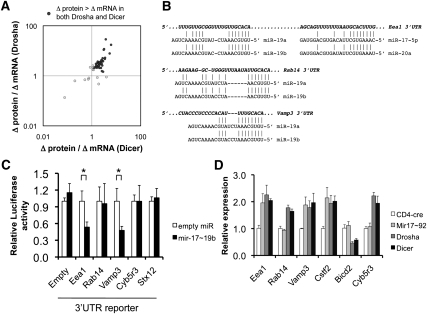
Figure 4.
Prediction of miRNA targets by measuring protein versus mRNA derepression in the absence of miRNAs. (A) Proteins that were up-regulated in both Drosha- and Dicer-deficient activated CD4+ T cells (from Fig. 3A) were analyzed for the magnitude of protein derepression versus mRNA derepression. The 3′UTRs of these genes were then analyzed for sites potentially targeted by miRNAs normally expressed in control cells. (B) Shown are predicted mir-17∼92a target sites in the 3′UTRs of Eea1, Rab14, and Vamp3, which were derepressed more at the protein than the mRNA level. (C) The entire 3′UTRs of Eea1, Rab14, and Vamp3 were inserted into a Firefly luciferase reporter and analyzed for reporter knockdown in the presence of overexpressed mir-17∼19b. (Mir-92a was left out of the expression construct because it was poorly processed.) Cyb5r3 and Stx12 3′UTRs were used as negative controls. The data represent the mean ± SEM of four experiments, normalized to Renilla luciferase as a transfection control. (*) P < 0.05 by t-test. (D) Confirmation of predicted mir-17∼92a targets in CD4+ T cells by specific deletion of the Mir17∼92a cluster. In vitro activated CD4+ T cells deficient in mir-17∼92a, Drosha, or Dicer were analyzed for expression levels of the predicted mir-17∼92a targets Eea1, Rab14, Vamp3, and Cstf2 by qRT–PCR. Analyses of Bicd2 and Cyb5r3 were included as negative controls. Bicd2, although a predicted mir-17∼92 target, was not derepressed in Drosha- or Dicer-deficient cells. The data represent the mean ± SEM of four experiments.