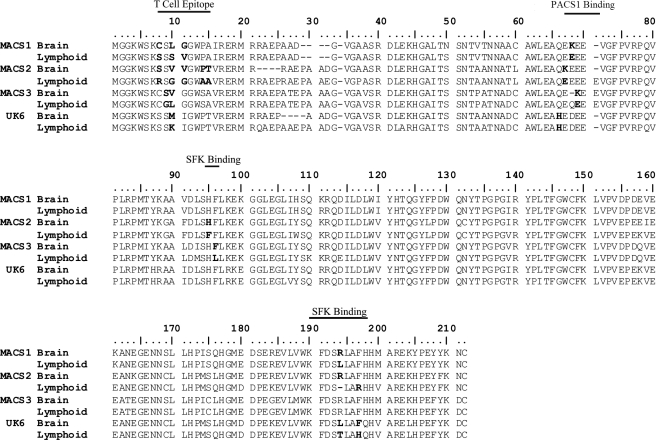
FIG. 2.
Alignment of predicted brain- and lymphoid-derived Nef amino acid sequences. Consensus nucleotide sequences were generated using Bioedit software from CLUSTAL W alignments of each patient-specific tissue set of sequences in which the representative base occurred in at least 60% of sequences in the set. Consensus nucleotide sequences associated with each patient-specific tissue were used to generate a predicted amino acid sequence. The resulting amino acid sequences were aligned with CLUSTAL W. Residues of interest are in bold. “T Cell Epitope” indicates residues within reported CTL or CD4+ T helper epitopes. “PACS1 Binding” indicates residues interacting with the PACS1 protein. “SFK Binding” indicates residues that may influence the association of Nef with Src family kinases.