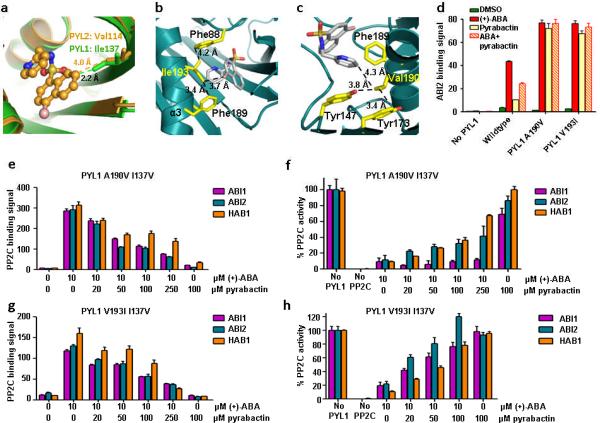
Figure 4.
I137V converts PYL1 into a pyrabactin-inhibited receptor. (a) PYL1 Ile137 clashes with the pyrabactin antagonist structure. Overlay of pyrabactin in the PYL2 ligand binding pocket (brown) with the PYL1 ligand binding pocket (green). Ile137 and the corresponding residue in PYL2, Val114, are represented as stick models. Their distances to pyrabactin in the PYL2 antagonist structure are indicated. (b,c) V193I and A190V mutations stabilize pyrabactin in the PYL1 pocket by forming hydrophobic interaction networks with pyrabactin and ligand binding pocket residues. (b) Model of Ile193 in the PYL1 pocket interacting with pyrabactin, Phe189, and Phe88, (c) Model of Val190 in the PYL1 pocket interacting with pyrabactin, Tyr173, Tyr147, and Phe189. (d) PYL1 A190V and V193I mutations increase the pyrabactin-mediated PYL1–ABI2 interaction. AlphaScreen interaction in the presence or absence of 10 μM (+)-ABA, 100 μM pyrabactin, or 5 μM (+)-ABA + 100 μM pyrabactin (n=3, error bars=s.d.). (e+g) Pyrabactin inhibits the ABA-stimulated interaction between PYL1 I137V and PP2Cs. Alphascreen interactions with PYL1 A190V I137V (e) or V193I I137V (g) (n=3, error bars=s.d.). (f+h) Pyrabactin relieves the ABA-stimulated inhibition of PP2C phosphatase activity by PYL1 A190V I137V (f) or V193I I137V (h) (n=3, error bars=s.d.).