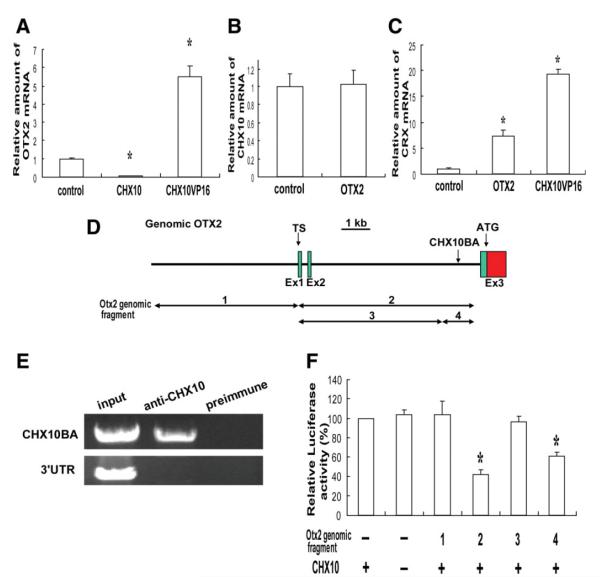
Figure 3.
Molecular interaction of CHX10, OTX2, and CRX in human retinal stem cell (hRSC) progeny. (A–C): Real-time reverse transcription polymerase chain reaction (RT-PCR) analysis. OTX2, CHX10, or CRX mRNA levels were normalized to GAPDH mRNA (the control sample was set to 1.0). (A): OTX2 mRNA level was significantly downregulated in CHX10-transduced hRSC progeny, whereas OTX2 mRNA was increased in CHX10VP16 transduced progeny (analysis of variance and Dunnette’s multiple comparison test, *p < .05). (B): CHX10 expression is not affected in OTX2 transduced hRSC progeny. (C): CRX mRNA levels were significantly increased in OTX2 or CHX10VP16-transduced hRSC progeny (analysis of variance and a Dunnette’s multiple comparison test, *p < .05). (D): Partial genomic map (5′ region) for the human OTX2 gene. Exons (Ex1,2, and 3), transcriptional start sites (TS) and the initiator codon (ATG) are indicated. A putative CHX10-binding area (CHX10BA) is located in intron2. (E): Chromatin immunoprecipitation analysis. Anti-Chx10 antibody specifically precipitates the chromatin containing the end of intron two of OTX2 (CHX10BA), but not the 3′UTR region (control), from hRSC progeny. Preimmune serum does not precipitate these regions. (F): Luciferase assay. OTX2 genomic fragments 1-4 shown in (D) were cloned into a luciferase reporter and cotransfected into hRSC progeny with or without the CHX10 expression vector. The activity of the βactin minimal promoter-luciferase reporter without genomic OTX2 was taken as 100%. Suppression of the reporter activity by CHX10 required the CHX10 binding area (CHX10BA) present in fragments 2 and 4 (analysis of variance and a Dunnette’s multiple comparison test, *p < .05). Abbreviations: TS, transcriptional start sites.