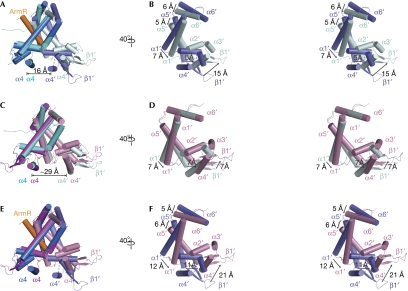
Figure 4.
Structural differences between ArmR-bound and oxidized MexR dimers. The positions of the DNA minor-groove-binding wing (β1′) and the helices, α1′, α2′, α3′, α4′, α5′ and α6′, in the unaligned protomers are labelled. All superimpositions were performed in PyMOL with one protomer aligned from each dimer. (A) Superimposition of ArmR-bound MexR and dimer CD with a core r.m.s.d. value of 2.8 Å2. The ArmR-bound MexR is shown with one protomer in marine and the other in slate, and the ArmR peptide in orange. The disordered loop between helices α3′ and α4′ in the ArmR-bound MexR is indicated by a dashed line. The distance between helices α4 and α4′ in the ArmR-bound MexR is 16 Å. (B) Stereoview of the unaligned protomer, as in (A), rotated 40° to the right. The conformational change of the DNA minor-groove-binding wing (β1′) and the dimerization helices, α1′, α5′ and α6′, is indicated with arrows. (C) Superimposition of the oxidized MexR and dimer CD to a core r.m.s.d. value of 1.6 Å2. The distance between helices α4 and α4′ in dimer CD is 29 Å. (D) Side view of the unaligned protomer as in (C) in stereo, rotated 40° to the right, revealing the conformational change due to the disulphide bonds formation. The conformational change of the DNA minor-groove-binding wing (β1′) and the dimerization helices, α1′ and α5′, is indicated with arrows. (E) Superimposition of the ArmR-bound and oxidized MexR dimers with a core r.m.s.d. value of 2.4 Å2. (F) Stereoview of the unaligned protomer as in (E), rotated 40° to the right, highlighting their significant conformational differences. The conformational change of the DNA minor-groove-binding wing (β1′) and the dimerization helices, α1′, α5′ and α6′, is indicated with arrows. cd, central distance.