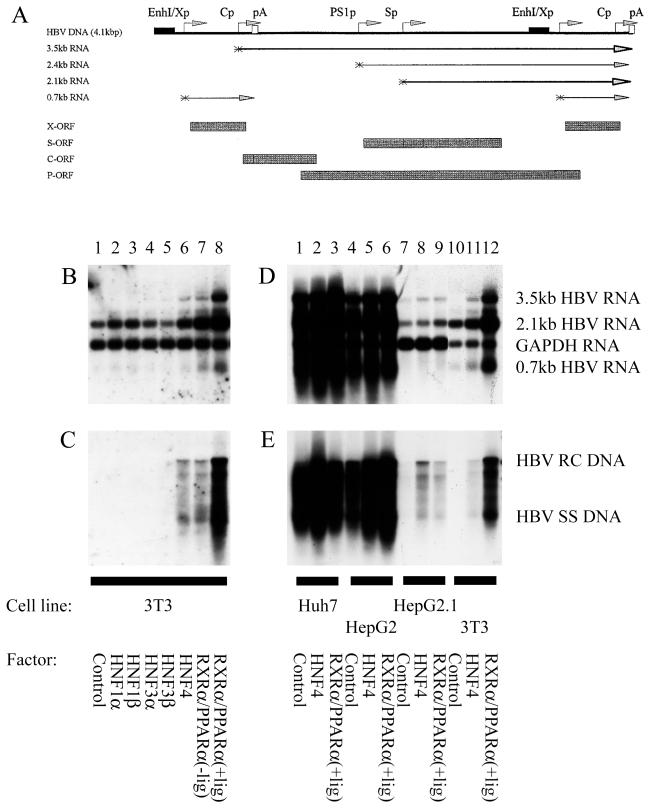
Figure 1.
Nuclear hormone receptors activate HBV replication in a nonhepatoma cell line. (A) Structure of the HBV DNA (4.1 kbp) construct used in transient transfection analysis. The 4.1-kbp greater-than-genome length HBV DNA sequence in this construct spans coordinates 1072–3182/1–1990 of the HBV genome (subtype ayw). The locations of the HBV 3.5-, 2.4-, 2.1-, and 0.7-kb transcripts are indicated. EnhI/Xp, enhancer I/X-gene promoter region; Cp, nucleocapsid or core promoter; pA, polyadenylation site; PS1p, presurface antigen promoter; Sp, major surface antigen promoter; X, X-gene; S, surface antigen gene; C, core gene; P, polymerase gene. (B–E) Cells were transiently transfected with the HBV DNA (4.1 kbp) construct and liver-enriched transcription factors. Mouse NIH 3T3 fibroblasts (3T3), human differentiated hepatoma cells (Huh7 and HepG2), and human dedifferentiated hepatoma cells (HepG2.1) were used for this analysis. (B and D) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (C and E) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. All-trans retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear hormone receptors RXRα and PPARα (+lig).