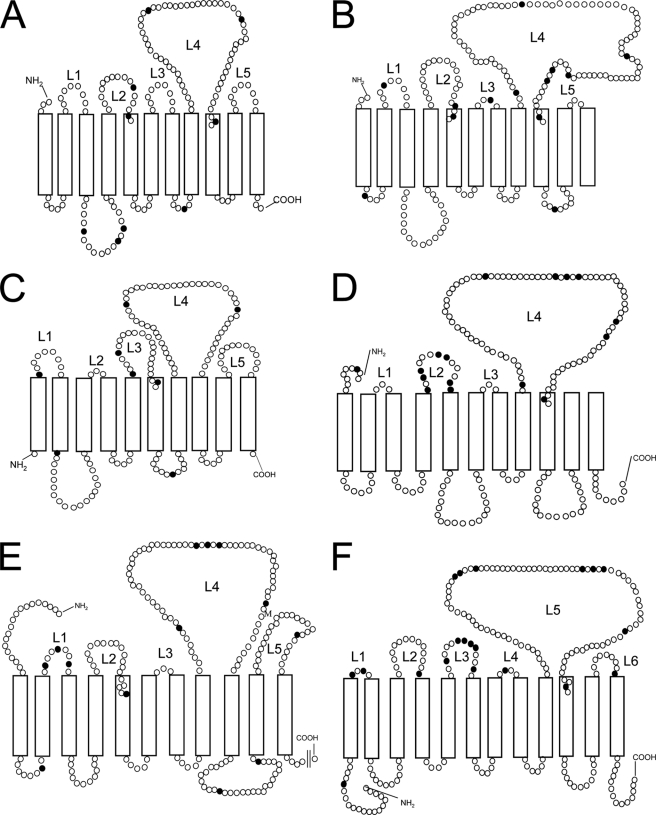
FIGURE 5.
Comparison of topology of O-antigen polymerase in several Gram-negative bacteria. Theoretical topology map of the O-antigen polymerases in E. coli O148 (A), Salmonella typhimurium LT2 (B), Bacteroides fragilis YCH46 (C), Pseudoalteromonas atlantica T6c (D), Marinobacter sp. ELB17 (E), and Thiomicrospira crunogena XCL-2 (F). Topology maps were predicted by TMHMM-2.0 and the previously published topology map of Shigella (33). Each circle represents an amino acid residue, and closed circles represent glycine residues. L1–L6, numbering of periplasmic loop (L).