FIGURE 8.
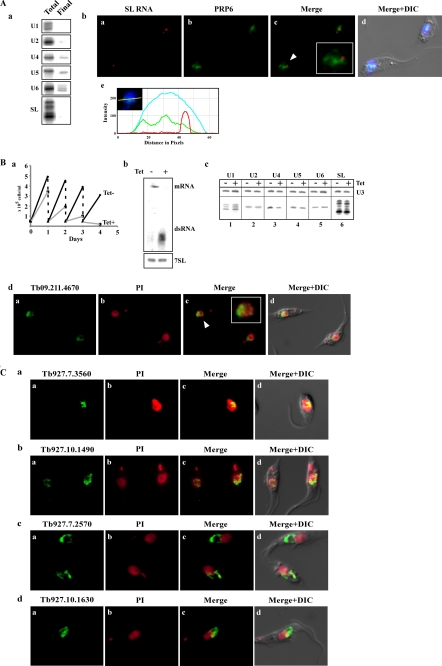
Localization of splicing factors co-purifying with SmD3 complex. A, PRP6. Panel a, PRP6 is associated with U5 but also with U4 and U6 snRNPs. Extract was prepared from cells as described (13). RNA was extracted from IgG-agarose beads and subjected to primer extension with the different probes (supplemental S-1). RNA was prepared from an aliquot of cells before selection (Total), and RNA from the beads (Final). Only an aliquot (2%) of the total RNA sample was analyzed, whereas the entire sample from the beads was analyzed. Panel b, localization of PRP6 with respect to SL RNA. In situ hybridization combined with immunofluorescence was performed as described under “Experimental Procedures.” Panel a, in situ hybridization with SL RNA; panel b, immunofluorescence of PTP-tagged PRP6; panel c, merge of panels a and b; panel d, DIC and DAPI-stained nuclei merge with panel c; panel e, intensity plot of the three chromophores under the line (inset) as described in Fig. 7D, panel e, demonstrating no significant overlap between the SL RNA and PRP6. B, gene coding for Tb09.211.4670 is essential and its silencing affects the level of U4 and U5 snRNAs. Panel a, growth curves of T. brucei cells silenced for the factor. Growth of uninduced cells was compared with growth after tetracycline addition. Both uninduced and induced cultures were diluted daily to 5 × 104 cells per ml. Panel b, Northern blot analysis demonstrating the silencing of Tb09.211.4670. RNA was prepared from uninduced cells (−Tet) and cells after 2 days of induction (+Tet). Total RNA (20 μg) was subjected to Northern analysis with randomly labeled probes. The transcripts of the gene, dsRNA as well as 7SL RNA, that were used to control for equal loading are indicated. Panel c, level of U snRNAs upon silencing. Total RNA as was extracted from uninduced cells or after 2 days of induction. The level of the U snRNAs was determined by primer extension using specific oligonucleotides as listed in supplemental S-1. U3 snoRNA level was used as a control for equal loading. Panel d, localization of the protein encoded by Tb09.211.4670. The factor was PTP-tagged, and the expression was detected by immunofluorescence. Panel a, immunofluorescence of PTP-tagged factor; panel b, propidium iodide-stained nuclei; panel c, merging of panels a and b; panel d, DIC merged with panel c. C, localization of the factors associated with SmD3. The details are the same as described in B, panel d, identity of each gene is indicated.