Abstract
Identification of clinical isolates of Neisseria meningitidis that are resistant to rifampin is important to avoid prophylaxis failure in contacts of patients, but it is hindered by the absence of a breakpoint for resistance, despite many efforts toward standardization. We examined a large number (n = 392) of clinical meningococcal isolates, spanning 25 years (1984 to 2009), that were collected in 11 European countries, Argentina, and the Central African Republic. The collection comprises all clinical isolates with MICs of ≥0.25 mg/liter (n = 161) received by the national reference laboratories for meningococci in the participating countries. Representative isolates displaying rifampin MICs of <0.25 mg/liter were also examined (n = 231). Typing of isolates was performed, and a 660-bp DNA fragment of the rpoB gene was sequenced. Sequences differing by at least one nucleotide were defined as unique rpoB alleles. The geometric mean of the MICs was calculated for isolates displaying the same allele. The clinical isolates displaying rifampin MICs of >1 mg/liter possessed rpoB alleles with nonsynonymous mutations at four critical amino acid residues, D542, H552, S548, and S557, that were absent in the alleles found in all isolates with MICs of ≤1 mg/liter. Rifampin-susceptible isolates could be defined as those with MICs of ≤1 mg/liter. The rpoB allele sequence and isolate data have been incorporated into the PubMLST Neisseria database (http://pubmlst.org/neisseria/). The rifampin-resistant isolates belonged to diverse genetic lineages and were associated with lower levels of bacteremia and inflammatory cytokines in mice. This biological cost may explain the lack of clonal expansion of these isolates.
Neisseria meningitidis is a strictly human bacterium encountered in the pharynx in about 10% of the general population (asymptomatic carriage) (31). This bacterium can also cause severe infections (mainly septicemia and meningitis) when it crosses the epithelial barrier to invade the bloodstream and the meninges (23). Close contacts of a patient with meningococcal disease have a highly elevated risk of contracting the disease. Preventive measures to avoid further spread and possible epidemics are recommended. These measures include the use of chemoprophylaxis and vaccination when isolates belong to one of the vaccine-preventable serogroups (A, C, Y, and W-135). In many European countries, rifampin is a first-line agent for chemoprophylaxis.
Several studies have reported rare N. meningitidis isolates that are considered resistant to rifampin. As in other bacterial species, this resistance is due mainly to mutations in the central part of the rpoB gene, encoding the β subunit of the RNA polymerase (6, 17). However, currently, this resistance seems to remain a rare event for N. meningitidis (20, 21), which may reflect a decreased biological fitness of rifampin-resistant N. meningitidis strains. Nevertheless, expansion of meningococcal clones resistant to rifampin may cause chemoprophylaxis failure, and reliable monitoring of rifampin susceptibility is crucial. Phenotypic determination of antibiotic susceptibility by use of an antibiogram is still not optimal, and there is a lack of correlation between laboratories, despite efforts toward technical standardization (30). Moreover, different breakpoints are employed by the national reference laboratories, and in addition, international committees such as the European Committee for Antimicrobial Susceptibility Testing (EUCAST) (http://www.srga.org/eucastwt/MICTAB/MICmiscellaneous.html) and the Clinical and Laboratory Standards Institute (CLSI) (8) recommend slightly different breakpoints. Rifampin-susceptible isolates are defined by rifampin MICs of ≤0.25 mg/liter and ≤0.5 mg/liter by the EUCAST and CLSI, respectively.
A recent study among members of the European Meningococcal Disease Society (formerly known as the EMGM) on penicillin G susceptibility in N. meningitidis proved that molecular characterization of a gene involved in resistance development can be used in addition to antibiograms in order to strengthen the categorization of susceptible and resistant isolates (25). This previous study was based on identification of alterations in the penA gene (encoding penicillin-binding protein 2, the target of penicillin). Critical alterations in penA that were linked directly to reduced susceptibility to penicillin G were confirmed by transformation of altered penA genes into a susceptible strain. Geometric means of MICs (MICgm) for isolates with altered penA alleles as well as wild-type penA alleles were then used to suggest the breakpoint for penicillin G susceptibility. In the present study, we aimed to apply this approach to rifampin in order to determine a breakpoint for rifampin in N. meningitidis by molecular analysis of the rpoB gene. We also aimed to address the impact of resistance mutations in the rpoB gene on the virulence of N. meningitidis isolates. The sequencing of the rpoB gene as well as the analysis of the impact of RpoB alterations on meningococcal virulence may help in understanding the biological cost of these alterations.
MATERIALS AND METHODS
Meningococcal isolates and phenotypic and genotypic characterization.
We included a large number (n = 392) of clinical N. meningitidis isolates that were collected between 1984 and 2009 in 11 European countries as well as in Argentina and the Central African Republic (Table 1). The collection comprised all clinical isolates with MICs of ≥0.25 mg/liter (n = 161) received by the national reference laboratories for meningococci in the participating European countries. In addition, representative isolates displaying rifampin MICs of <0.25 mg/liter were examined (n = 231). The latter isolates corresponded mainly to all French isolates received from December 2008 to January 2009. The isolates were mostly from invasive cases (n = 276 [70.4%]), with isolates from cerebrospinal fluid being the most frequent (40% of all isolates), followed by isolates from blood (26% of all isolates). The remaining isolates were obtained primarily from respiratory sites (mainly the nasopharynx). All isolates were identified using standard culture and identification methods. Serogroup was determined by agglutination with serogroup-specific antisera according to the standard procedure of each laboratory. Further phenotyping (serotyping and serosubtyping) was performed as previously described (1). Genotyping (using multilocus sequence typing [MLST], porA typing, and fetA typing) was performed as previously described (7, 9, 14, 22, 24, 28, 29). Sequence types (STs), clonal complexes (cc), and PorA and FetA types were determined through the meningococcal typing website (http://neisseria.org/nm/typing/).
TABLE 1.
Distribution of Neisseria meningitidis isolates according to rifampin MIC and country
| Country | No. of isolates with rifampin MIC (mg/liter) |
Total no. of isolates | |
|---|---|---|---|
| >1 | ≤1 | ||
| Argentina | 0 | 40 | 40 |
| Austria | 1 | 0 | 1 |
| Belgium | 1 | 9 | 10 |
| Central African Republic | 0 | 1 | 1 |
| Czech Republic | 0 | 36 | 36 |
| France | 11 | 139 | 150 |
| Germany | 0 | 34 | 34 |
| Greece | 1 | 3 | 4 |
| Italy | 10 | 0 | 10 |
| Poland | 1 | 22 | 23 |
| Romania | 0 | 19 | 19 |
| Spain | 4 | 38 | 42 |
| Sweden | 0 | 19 | 19 |
| Unknown | 3 | 0 | 3 |
| Total | 32 | 360 | 392 |
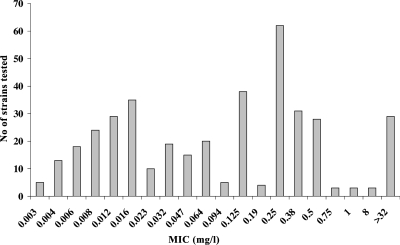
The MIC of rifampin was determined using the Etest method (AB Biodisk, Solna, Sweden) as previously described (30). The distribution of isolates according to their MIC values is shown in Fig. 1. The number of isolates per country and their exact MICs are available at the Neisseria PubMLST database (http://pubmlst.org/neisseria/) by filtering isolate queries with the dropdown project menu set to “rpoB sequence analysis.”
FIG. 1.
MICs (mg/liter) of rifampin in the examined meningococcal isolates (n = 392).
DNA sequencing and analysis of the rpoB gene.
The rpoB gene sequence for the sequenced strain (whole genome) MC58 (27) was used to design the following two primers for PCR amplification of a fragment of the rpoB gene (nucleotides 1256 to 2092), as previously described (26): RpoB1F, 5′-gttttcccagtcacgacgttgtaCTGTCCGAAGCCCAACAAAACTCTTGG-3′; and RpoB1R, 5′-ttgtgagcggataacaatttcTTCCAAGAATGGAATCAGGGATGCTGC-3′. Universal forward and reverse sequences were added as adapters for sequencing to the 5′ ends of the oligonucleotides (shown in lowercase letters). The universal forward and reverse sequences were then used for sequencing.
A DNA fragment of 660 bp of the rpoB gene, corresponding to amino acid residues 452 to 671 of the RpoB protein, was extracted from the DNA sequence. Alignments were made using the MacMolly program (Mologen, Berlin, Germany), and sequences differing by at least one nucleotide were assigned a unique rpoB allele sequence number. Phylogenetic networks were generated using Splits Tree 4.10 (www.splitstree.org) with default parameters (12).
Transformation of N. meningitidis.
rpoB amplicons from isolates with rifampin MICs of >32 mg/liter were used to transform the susceptible strain clone 12 as previously described, after purification on CL-6B Sepharose (Sigma Aldrich, Illkirch, France) (2). The rpoB amplicons were generated using the primers rpoB1UP (5′-ggccgtctgaaCTGTCCGAAGCCCAACAAAACTCTTGG-3′), containing the Neisseria-specific DNA uptake sequence (lowercase), and rpoB1R (11). Transformants were selected on GCB medium supplemented with Kellogg supplements (13) in the presence of rifampin at a concentration of 32 mg/liter. The rpoB gene was sequenced as described above to confirm the allelic replacement in the recipient strain by the transforming DNA.
Virulence analysis of rifampin-resistant isolates.
The impact of rpoB mutations in rifampin-resistant isolates on meningococcal virulence was evaluated by their ability to produce bacteremia in BALB/c transgenic mice expressing human transferrin (32). For this purpose, isogenic rifampin-resistant transformants harboring an alteration at residue S548 or S557 were compared to the susceptible parent strain clone 12 (the virulence of a transformant harboring the H552Y alteration has been examined previously) (26). Six-week-old female mice were infected with 0.5 ml of a standardized inoculum containing 5 × 106 CFU by the intraperitoneal route. Blood samples were taken after 6 h of infection, and CFU were determined by plating serial dilutions of blood on GCB medium. Blood samples were also used for quantification of cytokines (tumor necrosis factor alpha [TNF-α] and interleukin-6 [IL-6]) and of the chemokine KC (the murine homolog of human IL-8) by enzyme-linked immunosorbent assay (ELISA) (Quantikine; R&D Systems Europe, Abingdon, Oxon, United Kingdom). For competition experiments, we used our previously published rifampin-susceptible and rifampin-resistant isogenic strains (LNP21362 and NM05-08, respectively) (26). One volume (0.25 ml) of each strain containing 5 × 106 CFU was mixed and immediately injected by the intraperitoneal route into a 6-week-old female mouse. CFU were determined by plating on GCB medium of serial dilutions of the initial mixture (T0) as well as of blood samples that were taken after 2 and 6 h of infection. After 20 h of incubation, colonies were reisolated on plates of GCB medium and on plates of GCB medium in the presence of rifampin at a concentration of 32 mg/liter in order to determine the proportions of rifampin-susceptible and rifampin-resistant CFU. The experimental design was approved by the Institut Pasteur Review Board, Paris, France.
Statistical analysis.
Qualitative data were analyzed using the chi-square test. Statistically significant differences were assumed when P values were ≤0.05. Geometric means and lower and upper 95% confidence intervals were calculated using GraphPad InStat, version 3.06 (GraphPad Software, San Diego, CA). For reliable calculation of geometric means and 95% confidence intervals, only alleles that were represented by at least five isolates were included. The geometric mean was used because it better evaluates the “average” MIC for different isolates.
RESULTS
Characteristics of meningococcal isolates.
Serogroups were determined for 387 isolates (99%). Serogroup B (51%) was the most prevalent, followed by C (29%), W-135 (3%), Y (2.5%), and A (1.5%). Other serogroups or nongroupable isolates represented 13% of isolates, corresponding mainly to those recovered from respiratory sites. MLST was performed on 252 (64%) isolates representing 98 STs that were distributed among 22 distinct cc, while 12% (n = 31) of the isolates did not belong to any known cc and corresponded mainly to isolates from respiratory sites. Among strains with known STs, the five major hypervirulent cc in Europe were all represented: cc ST-11 (27%) was the most frequent, followed by cc ST-41/44 (18%), cc ST-32 (9%), cc ST-269 (6%), and cc ST-8 (5%). Other, less-frequent clonal complexes accounted for 23% of the isolates. Several FetA variants (n = 33) were observed among the 176 isolates (45%) for which FetA data were available. The most frequent variants were F1-5, F3-6, and F3-3, which accounted for 20%, 17%, and 15% of these isolates, respectively. Data on variable regions VR1 and VR2 of PorA were obtained for 264 isolates (67%), with 78 different combinations. The most frequent combination was VR1 type 5 with VR2 type 2, which accounted for 18% of the isolates with known data. All of these data underscore the heterogeneous aspect of the isolates of this study.
Characteristics of rpoB.
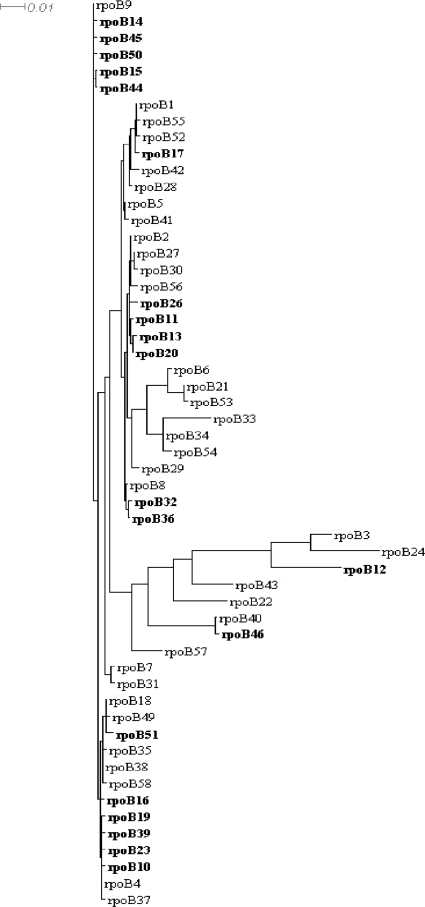
The sequence of a 660-bp DNA fragment of the central part of the rpoB gene was obtained for all 392 isolates included and allowed the identification of different rpoB alleles (n = 55) corresponding to 24 different RpoB amino acid sequences. The frequencies of these alleles varied from 1 to 98 isolates per allele. Phylogenetic relationships between alleles are shown in Fig. 2. All 55 rpoB alleles identified among the meningococcal clinical isolates in this study, as well as 3 other alleles detected in laboratory isolates or in other Neisseria species (data not shown), named rpoB1 to rpoB58, are available at the Neisseria meningitidis PubMLST sequence database (http://pubmlst.org/neisseria/). The most frequent allele among the examined isolates was rpoB4 (n = 98 isolates [25%]), which could be found in most of the participating countries, except for Argentina, where rpoB40 was the most frequent allele. The isolates harboring the rpoB4 allele were of several serogroups and STs, but serogroup C isolates belonging to cc ST-11 were most frequent (70%). When all alleles were aligned against the rpoB4 allele, they showed homologies ranging between 87.9% (80 polymorphic sites) and 99.9% (1 polymorphic site).
FIG. 2.
Schematic illustration of phylogenetic analysis of the 55 rpoB alleles that were identified among the tested isolates. Alleles that harbored modifications at residues S548, H552, and S557, as well as the D542V alteration, are shown in bold. The tree was generated using Splits Tree 4.10 (www.splitstree.org) with default parameters.
All isolates with MICs of >1 mg/liter (n = 32; MIC, 8 to 256 mg/liter) displayed nonsynonymous mutations at or close to amino acid residue H552 in RpoB and showed different phenotypes and genotypes (Table 2). However, two isolates with MICs of ≤1 mg/liter (0.125 mg/liter for both isolates) also contained amino acid alterations in this area (Table 2). At residue D542, only the D542V alteration was solely associated with a raised MIC and resistance to rifampin. Accordingly, the D542N alteration (rpoB57 allele), which was found in one isolate from Romania (MIC, 0.125 mg/liter) (Table 2), was not associated with resistance to rifampin unless it was accompanied by an alteration at another residue (e.g., rpoB26, with D542N and S557F mutations, found in one isolate from Greece, with a MIC of >32 mg/liter). Furthermore, amino acid alteration of residue G560 was not consistently associated with resistance to rifampin. For example, the rpoB37 allele, which harbors the G560S alteration, was associated with resistance to rifampin in two isolates from Spain (MIC, 8 mg/liter), while the same alteration did not render an elevated MIC in a French isolate (MIC, 0.125 mg/liter) that harbored the rpoB53 allele. These data suggest that alterations of three residues (S548, H552, and S557), as well as the D542V mutation, are directly involved or more involved than others in resistance to rifampin in N. meningitidis. The most frequent alteration was at residue H552 (n = 19 [56%]), which was replaced by Y, N, L, or R (Fig. 3 and Table 2). The corresponding isolates were from different serogroups and belonged to different genetic lineages.
TABLE 2.
Distribution of rpoB alleles with mutations at critical positions
| rpoB allele | No. of isolates | Country | MIC (mg/liter) | Amino acid alteration in RpoB | Serogroup:serotype: serosubtype | cc | PorA VR1 type | PorA VR2 type |
|---|---|---|---|---|---|---|---|---|
| rpoB10 | 1 | France | >32 | H552R | C:2a:P1.5,2 | 11 | 5 | 2 |
| rpoB11 | 1 | France | >32 | H552N | B:NT:NST | 32 | 19 | 15 |
| rpoB12 | 1 | France | >32 | H552L | B:15:P1.7 | 32 | 7 | 16-47 |
| rpoB13 | 1 | France | >32 | H552Y | B:15:P1.7 | 32 | 7 | 16-47 |
| rpoB14 | 1 | Unknown | >32 | S548F | ||||
| rpoB15 | 2 | Unknown | >32 | H552N | ||||
| rpoB16 | 2 | France | >32 | H552Y | C:NT:P1.6 | 269 | 18-1 | 3 |
| C:2a:P1.5 | ||||||||
| rpoB17 | 1 | France | >32 | H552Y | A:4:P1.9 | 5 | ||
| rpoB19 | 1 | France | >32 | S557F | C:2a:P1.5 | 11 | 5-1 | 10-8 |
| rpoB20 | 3 | France, Spain | >32 | H552Y | B:14:P1.7,16 | 32 | 7 | 16 |
| B:14:P1.7,16 | 32 | |||||||
| B:NT:P1.15 | ||||||||
| rpoB23 | 1 | France | >32 | S548F | C:2a:P1.7,1 | 11 | 7-1 | 1 |
| rpoB26 | 1 | Greece | >32 | D542N + S557F | B:15:P1.7,16 | 32 | 7 | 16 |
| rpoB32 | 1 | Poland | >32 | H552Y | W135:2a:NST | |||
| rpoB36 | 1 | Belgium | 8 | H552N | B:NT:P1.10 | UAa | ||
| rpoB37 | 2 | Spain | 8 | G560S | NG:NT:P1.3, B:NT:P1.3 | |||
| rpoB39 | 1 | Spain | >32 | D542V | B:2a:P1.5 | 11 | ||
| rpoB44 | 3 | Italy | 256 | H552Y | C:2b,4:P1.10 | 8 | 5-2 | 10 |
| rpoB45 | 3 | Italy | 256 | D542V | C:2b,4:P1.5 | 8 | 5 | 2 |
| C:2b,4:P1.5 | 8 | 5 | 2 | |||||
| B:2b,5:P1.10 | 8 | 5-2 | 10 | |||||
| rpoB46 | 1 | Italy | 256 | H552N | B:2b,4:P1.10 | 8 | 5-2 | 10 |
| rpoB50 | 3 | Italy | >32 | S557F | C:2a:P1.5 | 11 | 5 | 2 |
| rpoB51 | 1 | Austria | >32 | H552Y | ||||
| rpoB53 | 1 | France | 0.125 | G560S | B:NT:P1.4 | 41/44 | 7-2 | 4 |
| rpoB57 | 1 | Romania | 0.125 | D542N | B |
UA, unassigned.
FIG. 3.
Partial amino acid sequences of the central-terminal part of the β subunit of RNA polymerase (amino acids 538 to 563), deduced from the rpoB sequences of the N. meningitidis isolates examined. Apart from the rpoB4 sequence, only the sequences encoded by the rpoB alleles that harbored nonsynonymous mutations within this region are represented. Dots indicate identical residues, and polymorphic residues are indicated in one-letter code.
The mutations at residues S548, H552, and S557, as well as the D542V mutation, were absent in all rpoB alleles found in isolates with rifampin MICs of ≤1 mg/liter. The rpoB alleles that harbored mutations associated with high levels of rifampin resistance were distinct and differed from country to country. None of these alleles was detected in more than one country, except for the rpoB20 allele, which was detected in Spain and France (Table 2). In addition, no obvious correlation was observed between these alleles and resistance or reduced susceptibility to other antibiotics, such as ciprofloxacin or penicillin G (data not shown). In some of the participating countries (the Central African Republic, Argentina, the Czech Republic, Germany, Romania, and Sweden), rpoB alleles associated with rifampin resistance were not identified.
Correlation between MIC and alterations of the β subunit of the RNA polymerase.
To directly link mutations at residues S548, H552, and S557 to resistance to rifampin, a rifampin-susceptible strain, clone 12, was transformed with PCR products from the rpoB10, rpoB11, rpoB19, and rpoB23 alleles, harboring the amino acid alterations H552R, H552N, S557F, and S548F, respectively. Rifampin-resistant transformants (MICs, >32 mg/liter) were obtained with all four rpoB alleles, at a frequency of about 10−6 transformant/CFU. Sequence analysis of rpoB further confirmed that transformants harbored the same mutations as the transforming DNAs.
To further link susceptibility to rifampin to the absence of mutation at the S548, H552, and S557 residues (as well as the absence of the D542V mutation), the geometric means of the MICs (MICgm) of rifampin were calculated for the alleles without mutations at these positions. The two Spanish isolates harboring the rpoB37 allele were considered resistant strains through an as yet unidentified mechanism and were not included in the MICgm calculation. The MICgm for all of the isolates (n = 360) was 0.060 mg/liter (range, 0.003 to 1 mg/liter). The MICgm corresponding to alleles that were represented by at least five susceptible isolates varied from 0.025 to 0.225 mg/liter (n = 325) (Table 3). These data strongly suggest that the tested mutations are directly linked to meningococcal resistance to rifampin. Moreover, the absence of these mutations was strongly associated with low rifampin MICs (the highest MICgm was 0.225 mg/liter).
TABLE 3.
Distribution of rifampin MICs among the different rpoB alleles represented by at least five susceptible Neisseria meningitidis isolates (n = 325)
| rpoB allelea | No. of isolates | MIC range (mg/liter) | MICgm (mg/liter) | 95% Confidence interval (mg/liter) |
|---|---|---|---|---|
| rpoB1 | 32 | 0.047-0.5 | 0.225 | 0.173-0.294 |
| rpoB2 | 36 | 0.004-0.75 | 0.057 | 0.032-0.10 |
| rpoB4 | 98 | 0.003-1 | 0.089 | 0.066-0.12 |
| rpoB5 | 21 | 0.004-0.5 | 0.061 | 0.031-0.18 |
| rpoB9 | 27 | 0.003-0.75 | 0.065 | 0.033-0.128 |
| rpoB18 | 54 | 0.003-0.5 | 0.024 | 0.015-0.036 |
| rpoB27 | 8 | 0.015-0.5 | 0.15 | 0.052-0.43 |
| rpoB28 | 11 | 0.004-0.25 | 0.025 | 0.011-0.058 |
| rpoB31 | 11 | 0.004-0.06 | 0.015 | 0.008-0.027 |
| rpoB34 | 10 | 0.006-0.75 | 0.029 | 0.009-0.0.93 |
| rpoB40 | 17 | 0.008-0.12 | 0.034 | 0.023-0.051 |
Only alleles that were represented by at least five isolates are mentioned.
Impact of rpoB mutations on meningococcal virulence.
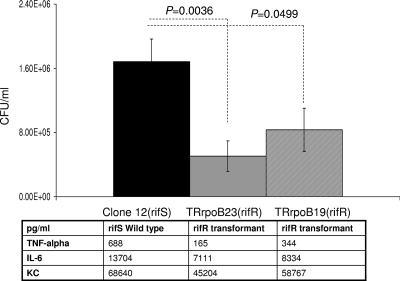
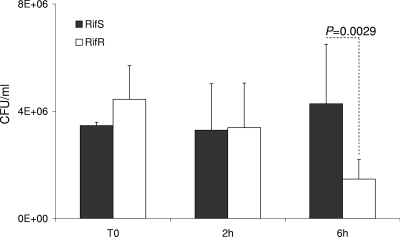
The two rifampin-resistant transformants, harboring the S548F (TRrpo23) and S557F (TRrpoB19) mutations, did not differ in growth rate from the wild-type parent strain when bacteria were cultured in liquid GCB medium (data not shown). To further investigate the impact of these mutations on meningococcal biology, we tested their effect on virulence. The ability of the rifampin-resistant transformants to provoke bacteremia in transgenic mice expressing human transferrin was compared to that of the wild-type strain. Significantly lower levels of septicemia (P < 0.05) were provoked by these transformants than by their isogenic parent strain (clone 12) (Fig. 4). Corroboratively, significantly (P < 0.05) lower levels of the inflammatory cytokines IL-6 and TNF-α and the chemokine KC (the murine homolog of human IL-8) were detected in the blood of mice infected by the two transformants than in that of mice infected by the wild-type strain (Fig. 4). All of these data suggest that rifampin-resistant isolates that result from mutations in the rpoB gene may show impaired virulence, as suggested by the lower levels of bacteremia in the experimental infections in mice. To further explore the impact of rpoB mutations on bacterial fitness during experimental infection, we also performed competitive infections, using our two previously described rifampin-susceptible and rifampin-resistant isogenic strains (LNP21362 and NM05-08, respectively) (26). Six-week-old mice (n = 10) were infected by intraperitoneal injection. The CFU number was determined for the initial mixture and for blood samples taken 2 and 6 h after infection. The numbers and proportions of rifampin-susceptible and rifampin-resistant colonies were determined by replicating the colonies in the presence or absence of rifampin as described in Materials and Methods. Bacterial counts for the rifampin-resistant strain were slightly higher at T0 than those for the susceptible strain. However, the counts of the resistant strain diminished and were significantly lower than those of the susceptible strain after 6 h of infection, representing only 25% of the whole bacterial population (Fig. 5). These data support the observation that rpoB mutations are associated with lower bacterial fitness during experimental infection.
FIG. 4.
Bacterial counts in blood from 6-week-old female transgenic BALB/c mice challenged intraperitoneally with standardized inocula of 5 × 106 CFU of Rifs (susceptible) isolates and two isogenic Rifr (resistant) transformants harboring the rpoB19 (S557F) or rpoB23 (S548F) allele. Results obtained after 6 h of infection are shown. The results shown are means for groups of at least five mice. P values were determined by two-tailed Student's t test. The levels of cytokines (TNF-α and IL-6) and of the chemokine KC are also displayed.
FIG. 5.
Bacterial counts in blood from 6-week-old female transgenic BALB/c mice. Animals were challenged by intraperitoneal injection with a bacterial mixture of two standardized inocula (about 5 ×106 CFU), using strain LNP21362 (Rifs) and its isogenic strain NM05-08 (Rifr), harboring the rpoB20 (H552Y) allele. Bacterial counts are shown for T0 and 2 and 6 h after infection. The results shown are means for groups of 10 mice for each point. P values were determined by two-tailed Student's t test.
DISCUSSION
Although meningococcal resistance to rifampin remains rare, it is important to effectively identify it because it may lead to failure of chemoprophylaxis in contacts of patients with meningococcal disease. Efforts have been made within the EMGM to standardize antibiotic testing techniques (e.g., the recommendation to use Etest on Mueller-Hinton agar supplemented with sheep blood) (30). However, establishment of harmonized, evidence-based breakpoints for resistance to rifampin remains crucial. This is highlighted by the fact that the breakpoints recently suggested by the EUCAST and the CLSI differ slightly and leave a range in which isolates can be misclassified.
The rpoB gene seems to show a certain extent of polymorphism that may be of interest in meningococcal typing. However, alterations of a limited number of positions in the rpoB gene have previously been suggested to be correlated with resistance to rifampin. In the present study, only three amino acid positions (S548, H552, and S557), as well as the D542V alteration, were identified as always and directly involved in conferring resistance to rifampin in N. meningitidis, in agreement with previous findings (17, 21). All isolates displaying mutations at any of the four critical positions of the rpoB gene displayed MICs of >1 mg/liter (Table 2). In addition, the MICgm of rifampin for the rpoB alleles without mutations at these positions was as low as 0.060 mg/liter (range, 0.003 to 1 mg/liter). The MICgm is a reliable tool for classifying isolates concerning their susceptibility to rifampin. In fact, the MICgm of a large collection of isolates sharing the same rpoB allele can help to indicate a reliable value for breakpoints because it smoothes out differences among laboratories in performing antibiotic susceptibility testing. Accordingly, based on the data from the present study, N. meningitidis isolates with MICs of ≤1 mg/liter should be considered susceptible to rifampin, since they lack the critical alterations of RpoB. It is noteworthy that this value is lower than the concentration of rifampin achieved in the epithelial lining fluid 2 to 5 h after a single dose of 600 mg of rifampin (33). Interestingly, our suggested breakpoint is identical to that proposed for rifampin on the basis of pharmacokinetic-pharmacodynamic (PK-PD) modeling with Monte Carlo simulation. Our biological data provide an experimental confirmation of the PK-PD assumption made for N. meningitidis (5).
One striking finding of this study is the high level of diversity among rifampin-resistant isolates. No clonal expansion of any “rifampin-resistant” allele could be detected, and the resistant isolates belonged to several diverse phenotypes and genotypes (Table 2). The “rifampin-resistant” alleles did not cluster together and were widely distributed in the phylogenetic tree, suggesting that they were derived from several “rifampin-susceptible” alleles (Fig. 2). This may be due to a defect in the ability of rifampin-resistant isolates to provoke invasive infections, i.e., to a decreased biological fitness upon invasiveness. Indeed, transmission rather than invasiveness derives selection of meningococci. We previously reported that rifampin-resistant isolates appeared most frequently as secondary cases upon use of rifampin in chemoprophylaxis. Rifampin-resistant isolates have also been obtained from carriers among contacts of an index case (20).
The data obtained from the virulence studies with the animal (transgenic mouse) model used in the present study, as well as a previous study on the role of the H552Y mutation (26), are in favor of a defect in the ability of these isolates to provoke septicemia. The level of bacteremia in mice is a good indicator of bacterial virulence, since it reflects bacterial survival upon invasion of the bloodstream. It is noteworthy that no growth defect was observed when bacteria were grown in liquid medium. Our data suggest that rifampin resistance in N. meningitidis may have a biological cost during infection in vivo. In Mycobacterium tuberculosis, rifampin resistance due to mutations in the rpoB gene leads to impaired fitness. Alterations in other genes (such as dnaE2) may then occur to restore fitness (4). Moreover, different mutations in rpoB may have different impacts on fitness through the survival rate of mutants, with subsequent differences in the frequency of the observed rpoB mutations (18). In Enterococcus faecium, the fitness cost of rifampin resistance due to mutation in rpoB is variable and can sometimes be absent (10). Mutations may also have different impacts according to their functional role in the RNA polymerase. For N. meningitidis, our data suggest that mutations at positions S548, H552, and S557 all reduce virulence in mice. Still, the impact of rpoB mutations on bacterial fitness requires further studies to explore the influence of these mutations on transcription during meningococcal infection.
The alterations of RpoB that confer rifampin resistance are located close to the active site of the RNA polymerase and the site of interaction with the σ subunit of the RNA polymerase, as well as to the tunnel through which the single-stranded DNA template must enter the RNA polymerase active site channel to base pair with the initiating nucleotide substrates (15, 16, 19). This may constrain the number of sites on the β subunit that can accept modification, and hence the small number of mutation sites in clinical isolates of N. meningitidis may result from specific structure requirements of the meningococcal RNA polymerase.
Three rpoB-related mechanisms have previously been reported to confer resistance to rifampin: (i) “steric” mutations (at amino acid residue S531 in Escherichia coli, corresponding to S557 in N. meningitidis), which exclude rifampin from interaction with RpoB; (ii) “affinity” mutations (at H526 in E. coli and H552 in N. meningitidis), which reduce antibiotic-β-subunit contacts; and (iii) “allosteric” mutations (at D516 in E. coli and D542 in N. meningitidis), which interfere with the transmission of the rifampin-induced allosteric signal and with allosteric modulation of the RNA polymerase catalytic reaction (3). Further experimental explorations are needed to study the impacts of these mutations on the function of RNA polymerase.
Residue G560 (mutated in the rpoB37 allele) in N. meningitidis has not been predicted to be involved directly in contact with rifampin (3), and accordingly, the G560S mutation was not consistently associated with resistance to rifampin. Moreover, we recently reported that transformation of a susceptible strain with a PCR fragment harboring this mutation did not confer resistance to rifampin (20). Other mechanisms may be responsible for the resistance to rifampin of the two Spanish isolates with the G560S mutation. An efflux system may be involved in rifampin resistance. However, we recently reported that there was no correlation between resistance to rifampin and alterations in the mtrR gene and its promoter (20). Finally, the impact of the S548F mutation on interaction with rifampin in N. meningitidis remains to be explored.
In conclusion, the data provided by the present study are valuable in establishing a molecular basis for both the phenotypic and genotypic surveillance of susceptibility to rifampin among N. meningitidis isolates. Based on the present findings, rifampin-susceptible isolates could be defined as those with MICs of ≤1 mg/liter and a lack of mutations at positions S548, H552, and S557, as well as a lack of the D542V alteration of rpoB, directly correlating with rifampin resistance. Alterations in these positions could also be correlated with reduced virulence of the bacteria in mice, but the impact of the rpoB mutations on bacterial fitness requires further studies to be understood fully.
Acknowledgments
Work on the isolates from Argentina was partially supported by a grant from the ECOS sud (A06S03). The study in Spain was partially supported by an FIS grant (PI060297) and by the Ministerio de Ciencia y Tecnologca, ISCIII, Spanish Network for Research in Infectious Diseases (REIPI RD06/0008).
Footnotes
Published ahead of print on 6 July 2010.
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1988. Neisseria meningitidis group B serosubtyping using monoclonal antibodies in whole-cell ELISA. Microb. Pathog. 4:27-32. [DOI] [PubMed] [Google Scholar]
- 2.Antignac, A., P. Kriz, G. Tzanakaki, J. M. Alonso, and M. K. Taha. 2001. Polymorphism of Neisseria meningitidis penA gene associated with reduced susceptibility to penicillin. J. Antimicrob. Chemother. 47:285-296. [DOI] [PubMed] [Google Scholar]
- 3.Artsimovitch, I., M. N. Vassylyeva, D. Svetlov, V. Svetlov, A. Perederina, N. Igarashi, N. Matsugaki, S. Wakatsuki, T. H. Tahirov, and D. G. Vassylyev. 2005. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell 122:351-363. [DOI] [PubMed] [Google Scholar]
- 4.Bergval, I. L., P. R. Klatser, A. R. Schuitema, L. Oskam, and R. M. Anthony. 2007. Specific mutations in the Mycobacterium tuberculosis rpoB gene are associated with increased dnaE2 expression. FEMS Microbiol. Lett. 275:338-343. [DOI] [PubMed] [Google Scholar]
- 5.Burgess, D. S., C. R. Frei, J. S. Lewis II, K. R. Fiebelkorn, and J. H. Jorgensen. 2007. The contribution of pharmacokinetic-pharmacodynamic modelling with Monte Carlo simulation to the development of susceptibility breakpoints for Neisseria meningitidis. Clin. Microbiol. Infect. 13:33-39. [DOI] [PubMed] [Google Scholar]
- 6.Carter, P. E., F. J. Abadi, D. E. Yakubu, and T. H. Pennington. 1994. Molecular characterization of rifampin-resistant Neisseria meningitidis. Antimicrob. Agents Chemother. 38:1256-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke, S. C., M. A. Diggle, and G. F. Edwards. 2001. Semiautomation of multilocus sequence typing for the characterization of clinical isolates of Neisseria meningitidis. J. Clin. Microbiol. 39:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Diggle, M. A., and S. C. Clarke. 2002. Rapid assignment of nucleotide sequence data to allele types for multi-locus sequence analysis (MLSA) of bacteria using an adapted database and modified alignment program. J. Mol. Microbiol. Biotechnol. 4:515-517. [PubMed] [Google Scholar]
- 10.Enne, V. I., A. A. Delsol, J. M. Roe, and P. M. Bennett. 2004. Rifampicin resistance and its fitness cost in Enterococcus faecium. J. Antimicrob. Chemother. 53:203-207. [DOI] [PubMed] [Google Scholar]
- 11.Goodman, S. D., and J. J. Scocca. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 85:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 13.Kellogg, D. S., Jr., W. L. Peacock, Jr., W. E. Deacon, L. Brown, and D. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami, K. S., S. Masuda, E. A. Campbell, O. Muzzin, and S. A. Darst. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285-1290. [DOI] [PubMed] [Google Scholar]
- 16.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 17.Nolte, O., M. Muller, S. Reitz, S. Ledig, I. Ehrhard, and H. G. Sonntag. 2003. Description of new mutations in the rpoB gene in rifampicin-resistant Neisseria meningitidis selected in vitro in a stepwise manner. J. Med. Microbiol. 52:1077-1081. [DOI] [PubMed] [Google Scholar]
- 18.O'Sullivan, D. M., T. D. McHugh, and S. H. Gillespie. 2005. Analysis of rpoB and pncA mutations in the published literature: an insight into the role of oxidative stress in Mycobacterium tuberculosis evolution? J. Antimicrob. Chemother. 55:674-679. [DOI] [PubMed] [Google Scholar]
- 19.Severinov, K., A. Mustaev, E. Severinova, M. Kozlov, S. A. Darst, and A. Goldfarb. 1995. The beta subunit Rif-cluster I is only angstroms away from the active center of Escherichia coli RNA polymerase. J. Biol. Chem. 270:29428-29432. [DOI] [PubMed] [Google Scholar]
- 20.Skoczynska, A., C. Ruckly, E. Hong, and M. K. Taha. 2009. Molecular characterization of resistance to rifampicin in clinical isolates of Neisseria meningitidis. Clin. Microbiol. Infect. 15:1178-1181. [DOI] [PubMed] [Google Scholar]
- 21.Stefanelli, P., C. Fazio, G. La Rosa, C. Marianelli, M. Muscillo, and P. Mastrantonio. 2001. Rifampicin-resistant meningococci causing invasive disease: detection of point mutations in the rpoB gene and molecular characterization of the strains. J. Antimicrob. Chemother. 47:219-222. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan, C. B., J. M. Jefferies, M. A. Diggle, and S. C. Clarke. 2006. Automation of MLST using third-generation liquid-handling technology. Mol. Biotechnol. 32:219-226. [DOI] [PubMed] [Google Scholar]
- 23.Taha, M. K., A. E. Deghmane, A. Antignac, M. L. Zarantonelli, M. Larribe, and J. M. Alonso. 2002. The duality of virulence and transmissibility in Neisseria meningitidis. Trends Microbiol. 10:376-382. [DOI] [PubMed] [Google Scholar]
- 24.Taha, M. K., D. Giorgini, M. Ducos-Galand, and J. M. Alonso. 2004. Continuing diversification of Neisseria meningitidis W135 as a primary cause of meningococcal disease after emergence of the serogroup in 2000. J. Clin. Microbiol. 42:4158-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taha, M. K., J. A. Vazquez, E. Hong, D. E. Bennett, S. Bertrand, S. Bukovski, M. T. Cafferkey, F. Carion, J. J. Christensen, M. Diggle, G. Edwards, R. Enriquez, C. Fazio, M. Frosch, S. Heuberger, S. Hoffmann, K. A. Jolley, M. Kadlubowski, A. Kechrid, K. Kesanopoulos, P. Kriz, L. Lambertsen, I. Levenet, M. Musilek, M. Paragi, A. Saguer, A. Skoczynska, P. Stefanelli, S. Thulin, G. Tzanakaki, M. Unemo, U. Vogel, and M. L. Zarantonelli. 2007. Target gene sequencing to characterize the penicillin G susceptibility of Neisseria meningitidis. Antimicrob. Agents Chemother. 51:2784-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taha, M. K., M. L. Zarantonelli, C. Ruckly, D. Giorgini, and J. M. Alonso. 2006. Rifampin-resistant Neisseria meningitidis. Emerg. Infect. Dis. 12:859-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, E. A., I. M. Feavers, and M. C. Maiden. 2003. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology 149:1849-1858. [DOI] [PubMed] [Google Scholar]
- 29.Urwin, R., J. E. Russell, E. A. Thompson, E. C. Holmes, I. M. Feavers, and M. C. Maiden. 2004. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect. Immun. 72:5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vazquez, J. A., L. Arreaza, C. Block, I. Ehrhard, S. J. Gray, S. Heuberger, S. Hoffmann, P. Kriz, P. Nicolas, P. Olcen, A. Skoczynska, L. Spanjaard, P. Stefanelli, M. K. Taha, and G. Tzanakaki. 2003. Interlaboratory comparison of agar dilution and Etest methods for determining the MICs of antibiotics used in management of Neisseria meningitidis infections. Antimicrob. Agents Chemother. 47:3430-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yazdankhah, S. P., and D. A. Caugant. 2004. Neisseria meningitidis: an overview of the carriage state. J. Med. Microbiol. 53:821-832. [DOI] [PubMed] [Google Scholar]
- 32.Zarantonelli, M. L., M. Szatanik, D. Giorgini, E. Hong, M. Huerre, F. Guillou, J. M. Alonso, and M. K. Taha. 2007. Transgenic mice expressing human transferrin as a model for meningococcal infection. Infect. Immun. 75:5609-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziglam, H. M., D. R. Baldwin, I. Daniels, J. M. Andrew, and R. G. Finch. 2002. Rifampicin concentrations in bronchial mucosa, epithelial lining fluid, alveolar macrophages and serum following a single 600 mg oral dose in patients undergoing fibre-optic bronchoscopy. J. Antimicrob. Chemother. 50:1011-1015. [DOI] [PubMed] [Google Scholar]